Caught in the Act: Cryo-EM Exposes γ-Secretase Catalytic Pose
Quick Links
A massive transmembrane complex, γ-secretase churns out the infamous peptides that can start a person on the path in Alzheimer’s disease. Aβ is produced when the secretase shaves three residues at a time from APP fragments within cell membranes. What controls this process? A cryo-EM study published June 7 in Science elucidates the mechanism.
- Cryo-EM resolves γ-secretase complexed with APP-C99, Aβ46, Aβ49, and Aβ43.
- During processing, an α-helix unwinds by one turn, while a three-residue β-strand positions it for the next cut.
- This mechanism aligns with the so-called “piston” model of γ-secretase processing.
Researchers led by Rui Zhou of Beijing’s Tsinghua University and Yigong Shi of Westlake University in Hangzhou resolved the structure of the secretase in complex with various Aβ peptides, including Aβ49, Aβ46, and Aβ43. The peptides lodged within a transmembrane channel of the enzyme, where each twisted into the same configuration: an α-helix connected via a short linker to a β-strand. For each successive cut, the substrate slid three residues, equivalent to one turn of the helix, deeper into the channel, while the C-terminus formed a hybrid β-strand with the enzyme that held the peptide in place for cleavage. In this manner, the secretase lops off three residues at a time.
“This new study is another tour de force in structure elucidation of the γ-secretase complex from [these investigators],” commented Michael Wolfe of the University of Kansas in Lawrence. “The snapshots of the protease bound to APP-derived [peptides] reveal critical details that, taken together, provide important insight into the mechanism of processive proteolysis.”
The study adds to prior work from Shi’s group that has brought γ-secretase into ever-clearer focus (Dec 2012 news; May 2015 news; Aug 2015 news).
Comprising the four subunits nicastrin, APH-1, PEN-2, and presenilin 1 or 2, the enzyme complex crisscrosses the cell membrane 20 times. Its catalytic heart beats within a channel formed by the sixth and seventh transmembrane domains of the presenilins. After β-secretase slices APP to leave a 99-amino-acid transmembrane fragment, aka β-CTF, γ-secretase goes to work. First using its endopeptidase activity, it clips C99 into either Aβ48 or Aβ49. Then, via processive, carboxypeptidase activity, it sequentially snips three to four residues from the C-terminus of those peptides. Depending on the endopeptidase cut, two production lines emerge: Aβ48 to 45 to 42, or Aβ49 to 46 to 43 to 40. Studies have demonstrated that familial AD mutations in the presenilins hobble both lines, leading to a glut of longer, more amyloidogenic peptides such as Aβ42 and Aβ43 and a dearth of Aβ38 and 40 (Apr 2019 news).
How does γ-secretase manage to precisely trim by three to four residues at a time? To find out, first author Xuefei Guo and colleagues resolved the structure of the enzyme in complex with peptides of the Aβ49 production line, including APP-C99, Aβ49, Aβ46, and Aβ43. To capture each substrate in the clutches of the enzyme, the researchers used trial and error to introduce cysteine residues in each peptide, and in the nicastrin subunit, that would allow them to cross-link the pair without distorting the substrate in the active site. Nicastrin helps capture and deliver substrates to the catalytic core of γ-secretase (Aug 2015 news; Dec 2015 news). To hold the pair in an intermediate state, the researchers introduced a catalysis-quashing D385N mutation into PS1’s catalytic site.
First, the scientists resolved the C99 complex at 2.9 Å resolution. They spied residues 31-55 to be nestled within the catalytic transmembrane channel of PS1. The segment’s N-terminus had an α-helix that lay next to a three-residue linker sequence, connected to a five-residue β-strand at the C-terminus. Hydrogen bonds between the α-helix and PS1’s transmembrane strand held the substrate in place. Meanwhile, the C99 β-strand coupled with two β-strands from PS1 to position the scissile peptide bond, aka the bond about to be snipped, in the linker between residues 49 and 50. This would produce Aβ49 and C50-99, aka the APP intracellular domain, or AICD.

First Cut. A bit of APP-C99 (orange) lodges into the transmembrane catalytic channel of PS1 (blue, left). The substrate’s α-helix connect to a β-strand via a linker (middle). This β-strand forms a hybrid with PS1 (close-up, right). [Courtesy of Guo et al., Science, 2024.]
To see how the structure then changes to accommodate the next, carboxypeptidase cut, the scientists next resolved the Aβ49 complex. Lo and behold, a similar scene emerged. A segment of Aβ49 formed an α-helix connected to a C-terminal β-strand. Relative to the positioning of C99, the Aβ49 helix was buried by three amino acids, or one turn of the helix, deeper into the transmembrane cavity of PS1. It was held in place by hydrogen bonding with the same two residues from PS1 that had cradled C99’s helix. The linker and β-strand shifted by three residues as well, such that the three residues that had formed the linker of C99 now folded into a β-strand in Aβ49, and the five residues that had folded into a β-strand in C99 were released from the enzyme’s grip. Aβ49’s short β-strand hooked up with one β-strand from PS1, not two. This positioning poised the substrate for cleavage between V46 and I47, which would yield Aβ46.
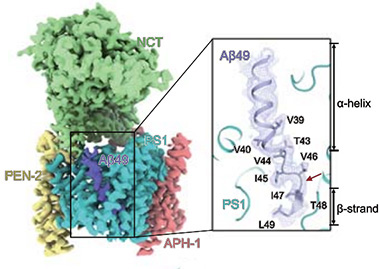
Next on the Block. In the clutches of γ-secretase (left), Aβ49 (purple) forms an α-helix (right) and a three-residue β-strand that pairs with PS1 to position the peptide for a chop after residue 46 (red arrow). The helix slides one turn deeper into the catalytic cleft relative to the position of the C99 helix. [Courtesy of Guo et al., Science, 2024.]
You guessed it: A nearly identical structure emerged in the Aβ46 complex. An N-terminal α-helix, three amino acids deeper into the catalytic cleft, was held in place by the same two residues of PS1, while a new C-terminal three-residue β-strand formed a hybrid β-sheet with PS1. With the bond between T43 and V44 now splayed across the PS1 chopping block, the enzyme was primed to produce Aβ43.
Try as they might, the researchers were unable to resolve an Aβ43 peptide complexed with γ-secretase at atomic resolution. However, at 4.45Å resolution, they made out the characteristic α-helix and hybrid β-strand structure, suggesting Aβ43 was likely cleaved in the same way as its predecessors.
The findings support the so-called “piston” model of γ-secretase processing, whereby the whole substrate edges closer to the jaws of the PS1 catalytic site after each cleavage (Jan 2019 news). The size of each bite is dictated by the pitch of the α-helix, which is three amino acids long. The authors think the enzyme likely works this way for the other Aβ production line, leading from Aβ48 to Aβ42, as well as for other γ-secretase substrates, such as Notch-1.
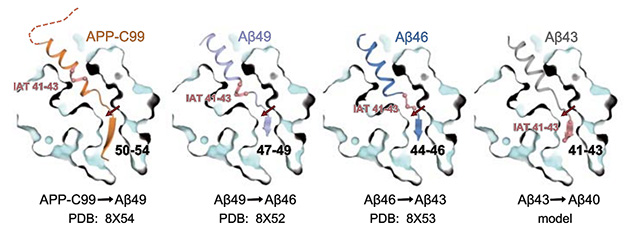
One Turn at a Time. In the proposed model, each successively smaller APP fragment forms an α-helix paired with a short β-strand that holds the substrate in place for cleavage. Bolded numbers indicate β-strand residues for each structure. The tripeptide IAT (pink, amino acids 41-43) is shown for reference; it moves closer to the active site (red arrow) with each cut. [Courtesy of Guo et al., Science, 2024.]
“The adjustment of the helical region of bound Aβ intermediates—into the hydrophobic presenilin cavity and toward the active site to set up tripeptide trimming—offers incontrovertible proof for the ‘piston’ model of processivity,” noted Wolfe. That said, he likened it to a ratchet rather than a piston, because the helical region only moves in one direction, not back and forth.
Of course, given its reliance on a series of static structures, cryo-EM cannot address exactly how this racheting takes place, Wolfe noted. “Conformational adjustments of the enzyme might involve fleeting dissociation of presenilin loop 1 from the substrate, and widening of the presenilin hydrophobic channel and active site,” he proposed.
Guo and colleagues’ reported structures come on the heels of another cryo-EM analysis of γ-secretase, led by Lucia Chavez-Gutierrez and Rouslan Efremov of KU Leuven in Belgium. These scientists elucidated γ-secretase in complex with the Aβ46 peptide. Their 27 May Nature Communications paper focused on how interactions between the B isoform of AphI and PS1 subunits might lead to allosteric regulation of proteolysis. With regard to how the enzyme latches onto Aβ, the two papers report somewhat different findings. For example, first author Ivica Odorčić and colleagues resolved an unstructured segment resting upon loop-1 of PS1, suggesting that “a close interaction with this loop reduces the flexibility of this substrate region and makes it detectable in cryo-EM,” Odorčić and Chávez Gutiérrez wrote to Alzforum. Guo’s study did not detect this interaction. “Its functional significance is unclear, but the presence of Alzheimer’s-disease-linked mutations in this loop support its involvement in efficient enzyme processivity,” they wrote (comment below).
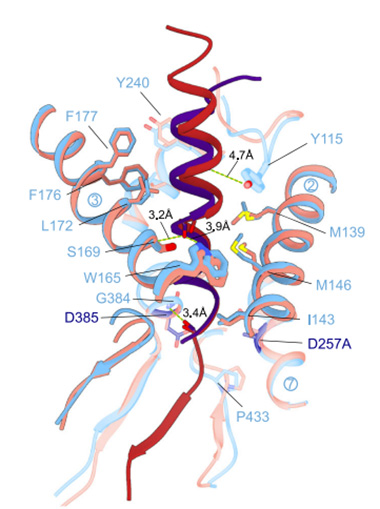
A Familiar Fold. In complex with PS1 (pink/light blue), both Aβ46 (red) and APP-C83 (dark blue) form an α-helix connected to a β-strand. Hydrogen bonds connect PS1 to its catalytic prey. [Courtesy of Odorčić et al., Nature Communications, 2024.]
Important similarities between the papers were the β-sheet at the C-terminal end of the substrate that formed a hybrid with PS1, and hydrogen bonds between residues Tyr115, Ser169 and Trp165 in PS1 and the substrate α-helix (image at right). Like Guo and colleagues, Odorčić found the β-strand was longer in C83, before the endopeptidase cut, than in Aβ46 but, other than that, the structure was maintained. “Collectively, these data show that the substrate conformation is largely similar between initial and intermediate cuts, suggesting that GSEC shapes the substrate as the substrate rearranges during processive proteolysis at its C-terminus,” the authors wrote.
Odorčić et al. also reported that eliminating the helical hydrogen bonds substantially hobbled the processive capacity of the enzyme, resulting in longer Aβ peptides. “Disruption of these bonds, through mutations, likely explains the pathogenicity of early onset Alzheimer’s disease variants at these positions,” Odorčić and Chávez Gutiérrez wrote.—Jessica Shugart
References
News Citations
- First Crystal Structure of a Presenilin
- Gamma Secretase: Intramembrane Liaisons Revealed
- γ-Secretase Revealed in Atomic Glory
- Familial Alzheimer’s Mutations: Different Mechanisms, Same End Result
- Nicastrin Bounces Bulky Proteins from γ-Secretase
- CryoEM γ-Secretase Structures Nail APP, Notch Binding
Mutation Interactive Images Citations
Further Reading
No Available Further Reading
Primary Papers
- Guo X, Li H, Yan C, Lei J, Zhou R, Shi Y. Molecular mechanism of substrate recognition and cleavage by human γ-secretase. Science. 2024 Jun 7;384(6700):1091-1095. Epub 2024 Jun 6 PubMed.
- Odorčić I, Hamed MB, Lismont S, Chávez-Gutiérrez L, Efremov RG. Apo and Aβ46-bound γ-secretase structures provide insights into amyloid-β processing by the APH-1B isoform. Nat Commun. 2024 May 27;15(1):4479. PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.