In Lipoparticles, ApoE Double Belt Keeps the Fat In
Quick Links
The structure of ApoE has proven tricky to discern in its native state, namely, attached to lipoproteins. Now, scientists led by David Holtzman at Washington University in St. Louis have captured lipidated ApoE at its highest resolution yet. In a first, they saw it bound to brain-derived lipoproteins. As reported in the January 23 Neuron, transmission electron microscope (TEM) images show two ApoEs circling disc-shaped lipoprotein particles isolated from the mouse brain, confirming previous images of artificial liposomes.
- Cryo-EM spies ApoE hugging lipoproteins in a “double belt” structure.
- Two antiparallel α-helical chains wrap around lipoprotein discs.
- Individual helices were not resolved.
Cryo-EM brought the complexes into slightly sharper focus, supporting the idea that α-helical chains of ApoE wrap in opposite directions around the discs, like frosting piped around the side of a cake. Still, at a resolution of 8 Å, individual helices could not be mapped.
“The results … are consistent with the antiparallel ‘double-belt’ model of ApoE [and] provide strong supportive evidence for [it] but do not directly prove it due to limited resolution,” wrote Olga Gursky of Boston University (comment below).
Scientists have struggled to elucidate the structure of lipidated ApoE with biophysical techniques because, even when crystallized or frozen, the proteins shimmy slightly within lipid bilayers. Previously, scientists led by Karl Weisgraber, University of California, San Francisco, used X-ray crystallography to determine the structure to 10 Å. This suggested that two ApoE proteins bent into horseshoe shapes that partially circled the lipid particles (Peters-Libeu et al., 2006). Based on cysteine cross-linking analysis, Vasanthy Narayanaswami of California State University in Long Beach and colleagues proposed that two ApoE proteins wrap lipids or lipoproteins in antiparallel or hairpin conformations (Kothari et al., 2021).
To capture a higher-resolution image, first author Michael Strickland cultured astrocytes, the primary source of the apolipoprotein in the brain, from mice expressing human APOE2, APOE3, or APOE4, then isolated lipoproteins from the cell media. He first analyzed them using negative-stain transmission electron microscopy (TEM), which keeps ApoE in its native conformation. In this method, heavy metal ions tint the glass slide dark while leaving the lipid particles untouched and bright under the microscope, enabling Strickland to view the particle's size and morphology at a resolution of 20 Å.

Fuzzy but Fab. A TEM image shows the HJ15.30 N-terminal ApoE Fab binding a lipoprotein particle between two HJ15.10 mid-region antibodies. The pattern supports the double-belt model of ApoE binding. (Courtesy of Strickland et al., Neuron, 2024.)
Strickland added antibodies to partially immobilize ApoE and help orient it on the lipoprotein particles for TEM. He used two different Fab antibody fragments. HJ15.30 binds residues 40-70 near ApoE's N-terminus, and HJ15.10 binds amino acids 140-160 in its middle. Two molecules of HJ15.10 latched onto almost opposite sides of each particle, suggesting that two ApoEs bound with their midsections facing away from one another. One bivalent HJ15.30 Fab fragment attached between the two other antibodies, suggesting it bound both N-termini and that they had to be adjacent (image below).
To the authors, this means that a pair of ApoE proteins stretched in opposite directions around the lipoprotein. “There’s no way the antibodies can line up that way unless ApoE forms an antiparallel dimer,” Holtzman told Alzforum.
The same arrangement emerged when the scientists generated lipoprotein particles by mixing recombinant ApoE with a phosphocholine derivative. All three ApoE isoforms, be they recombinant or from mouse astrocytes, bound lipids in the same pattern.
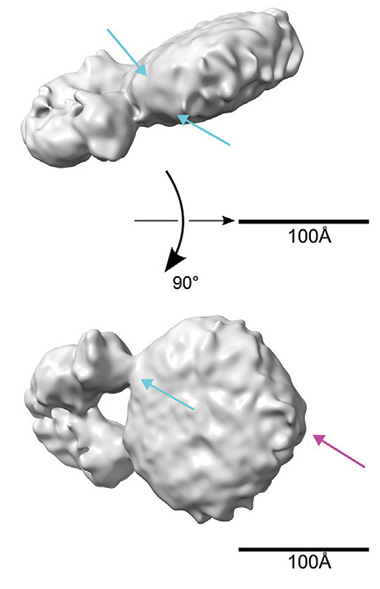
Toward 3D ApoE. A cryo-EM density map suggests that the HJ15.30 antibody (Y-shape jutting out from the disc) straddles two ApoEs. Blue arrows point to ridges of electron density believed to be the ApoE N-terminals. The fuchsia arrow points to what might be a kink in the helical chain, possibly the hinge region of ApoE between the N- and C-termini. (Courtesy of Strickland et al., Neuron, 2024.)
To get a clearer picture, Strickland turned to cryo-EM of lipoproteins prepared in vitro. Again, he used the antibody Fab’s. Both latched to the lipoprotein discs just as they had in the TEM images. When the scientists mapped electron density in three dimensions, they were able to vaguely make out two ridges along the circumference of the lipoprotein discs, consistent with the ApoE double-belt concept. By straddling both proteins, the HJ15.30 Fab appeared to lock them together and minimize their squirming. This allowed the scientist to resolve the structure to 8 Å, Strickland speculated (image below).
To the authors, the density map suggests that ApoE was wrapped around the perimeter, like two ribbons around the cream in an Oreo cookie (see model below). They think ApoE is likely extended in an α-helix chain, with the hydrophobic residues tucked into the fatty acid chains of the lipid particles and the hydrophilic residues residing in the solvent. This shape has been proposed for how ApoA1, ApoE's cousin, binds high-density lipoproteins in the blood (Wlodawer et al., 1979; reviewed by Gurksy, 2005; Bedi et al., 2022).
Narayanaswami agreed that the two ApoEs might be antiparallel, but noted that a hairpin conformation also explains the data. Rather than two ApoEs stretching around the lipid bilayer as “belts,” each forms a hairpin that wraps around half of the lipoprotein with the ApoE N-termini lying head-to-head. This would give a similar antibody binding pattern.
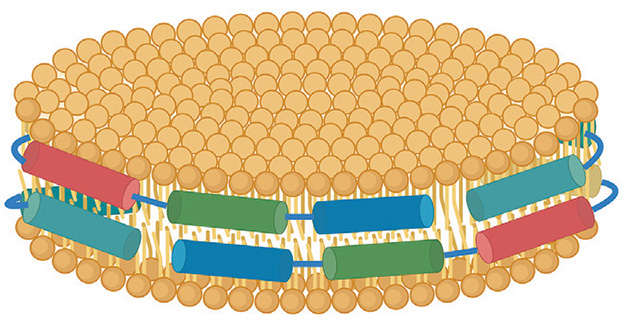
Keeping It All In. The proposed model of lipidated ApoE has two antiparallel α-helical chains (cylinders) wrapped around a lipid disc (yellow circles). [Courtesy of Strickland et al., Neuron, 2024.]
Others were surprised that, at a resolution of 8 Å, the structure lacked detail. “The protein position on the lipid is not clearly discernable [and] one should be able to resolve individual α-helices in the double belt since the helical diameter is over 10 Å,” wrote Gursky. Sjors Scheres of the Medical Research Council Laboratory of Molecular Biology at Cambridge, U.K., agreed. “Despite a statement in the text that the authors see densities for the α-helices, the map deposited in the database does not show the expected tubular densities one would expect,” he wrote to Alzforum.
Still, these cryo-EM images are a step toward understanding lipoprotein particle structure. “This study paves the way for obtaining high-resolution structural information of lipidated ApoE,” wrote Guojun Bu, Hong Kong University of Science and Technology, China (comment below). What will get researchers there all the way? “I think it’s going to take another technological advance in either cryo-EM or another method to get to 3 Å resolution,” Holtzman said.—Chelsea Weidman Burke
References
Mutation Interactive Images Citations
Research Models Citations
Paper Citations
- Peters-Libeu CA, Newhouse Y, Hatters DM, Weisgraber KH. Model of biologically active apolipoprotein E bound to dipalmitoylphosphatidylcholine. J Biol Chem. 2006 Jan 13;281(2):1073-9. Epub 2005 Nov 8 PubMed.
- Kothari S, Bala N, Patel AB, Donovan A, Narayanaswami V. The LDL receptor binding domain of apolipoprotein E directs the relative orientation of its C-terminal segment in reconstituted nascent HDL. Biochim Biophys Acta Biomembr. 2021 Jul 1;1863(7):183618. Epub 2021 Apr 6 PubMed.
- Wlodawer A, Segrest JP, Chung BH, Chiovetti R Jr, Weinstein JN. High-density lipoprotein recombinants: evidence for a bicycle tire micelle structure obtained by neutron scattering and electron microscopy. FEBS Lett. 1979 Aug 15;104(2):231-5. PubMed.
- Gursky O. Apolipoprotein structure and dynamics. Curr Opin Lipidol. 2005 Jun;16(3):287-94. PubMed.
- Bedi S, Morris J, Shah A, Hart RC, Jerome WG, Aller SG, Tang C, Vaisar T, Bornfeldt KE, Segrest JP, Heinecke JW, Davidson WS. Conformational flexibility of apolipoprotein A-I amino- and carboxy-termini is necessary for lipid binding but not cholesterol efflux. J Lipid Res. 2022 Mar;63(3):100168. Epub 2022 Jan 17 PubMed.
Further Reading
No Available Further Reading
Primary Papers
- Strickland MR, Rau MJ, Summers B, Basore K, Wulf J 2nd, Jiang H, Chen Y, Ulrich JD, Randolph GJ, Zhang R, Fitzpatrick JA, Cashikar AG, Holtzman DM. Apolipoprotein E secreted by astrocytes forms antiparallel dimers in discoidal lipoproteins. Neuron. 2024 Apr 3;112(7):1100-1109.e5. Epub 2024 Jan 23 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.