Single-Cell Profiling Maps Human Microglial Diversity, Flexibility
Quick Links
A first go at cataloging gene expression in individual microglia from human brain reveals a picture similar to that seen in mice: Microglia come in many flavors, morphing over time with age and brain location, and in response to disease. In identifying major subtypes of human microglia, researchers led by Marco Prinz, University of Freiburg, Germany, provide critical baseline data for future studies of neurodegenerative disease, and some reassurance that mice can replicate human response, at least where microglia are concerned. The work appeared February 13 in Nature.
- Scientists report first single-cell RNAseq comparison of human and mouse microglia.
- Cells are temporally and spatially diverse in development and disease.
- Similarities among mouse and human microglia bode well for models.
As the first responders of the innate immune system in the brain, microglia are proving to be central to Alzheimer’s disease pathogenesis and progression (Feb 2019 news). Many genetic risk factors for AD influence microglia function. In terms of new treatments, the cells are an attractive but moving target, since they readily adopt different phenotypes. To better pin down these diverse and dynamic players, researchers are moving from transcriptomic analysis of cells in bulk to single-cell RNA sequencing.
In the new study, first authors Takahiro Masuda, Roman Sankowski, and Ori Staszewksi started out by isolating and profiling microglia from mice. After sorting and sequencing, they clustered the cells based on similarity of gene expression. In analyzing a total of 3,826 single microglia from multiple brain regions of embryonic, juvenile, and adult mice, the researchers identified six distinct microglia subsets unique to the developing mouse brain. Once mice matured, most microglia assumed a more uniform, homeostatic profile. That state changed when they were challenged. Facial nerve injury, which triggers acute activation of microglia in the cortex and neurodegeneration, induced a novel cluster within three days of injury, which was gone two weeks later. Feeding the mice cuprizone, which causes reversible demyelination of axon tracts in the corpus callosum, induced two other injury-specific cell phenotypes over time, one during the demyelination period, and the other coinciding with remyelination.
The results mirror two recent single-cell studies on mouse microglia and bear out earlier speculation that the cells respond with lesion-specific changes in gene expression (Dec 2018 news; Li et al., 2109).
“This is important work, and shows that there is not just one phenotype of disease-related microglia, but multiple phenotypes,” said Oleg Butovsky, Brigham and Women’s Hospital, Boston. The field must move beyond a simple concept of disease-associated microglia, or neurodegeneration-associated microglia that have been described by his lab and others, he said. “One day we will have names for as many microglia subsets as there are T cells,” Butovsky speculated.
What about human cells? Compared with their murine counterparts, live human microglia are harder to come by. For this work, Prinz and colleagues isolated microglia from healthy cortical tissue surgically removed from five people with epilepsy, who averaged 42 years old. In addition, they acquired diseased cortical tissue from five cases of early multiple sclerosis, mean age of 36, who had undergone diagnostic biopsies of active lesions. In total, the researchers analyzed 1,602 microglia.
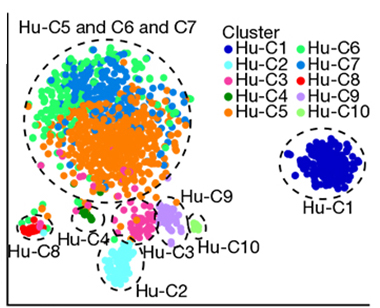
Clustering. Subtypes of human microglia were defined by transcriptional profiling. Each dot represents one cell, colors show cells belonging to each cluster. Clusters C5, C6, and C7 are homeostatic microglia; C4 comprises partially activated cells; C2, 3, and 8 are detected in MS lesions. C1, 9, and 10 are non-microglia cell types. [Courtesy of Masuda et al., 2019 Nature.]
Clustering by gene expression levels yielded seven distinct clusters of human microglia. Three contained cells exclusively from the healthy tissue samples. These expressed high levels of core microglia genes, consistent with homeostatic microglia. A fourth cluster, present in both healthy and MS brains, had reduced expression of core genes such as TMEM119 and upregulation of chemokine and cytokine genes involved in the innate immune response, including CCL2, CCL4, and EGR2. These were likely partially or pre-activated microglia, the authors decided. Finally, they found two additional clusters enriched in cells from MS brain, and one unique to MS brain, all of which clearly differed from the homeostatic microglia. These MS clusters had little or no TMEM119, and upregulated APOE and genes related to immune function and regulation. Further, while the mix of homeostatic microglia subtypes differed from person to person, the disease-associated cells varied little among patients. “This was a major surprise for us and very exciting,” said Prinz. “These early active cases have similar microglia subsets that form a kind of microglia signature in MS brain. These are the cells we might target with new treatments,” he said.
The analysis yielded a bonus—identification of expression markers for each subtype. Through immunostaining of up to four of these protein markers at a time, the researchers identified microglia in intact tissue. Richard Ransohoff of Third Rock Ventures, Boston, praised their efforts. “Using more than one or two markers to discriminate the different cells in tissues is exemplary, and is the way it should always be done,” Ransohoff said. “This is going to be a fantastic data resource. Their hard work and tremendous rigor are going to pay off,” he told Alzforum.
Faithful recapitulation of human immune responses remains a concern when using animal models. However, in a direct comparison, the human and mouse microglia phenotypes correlated surprisingly well, Prinz said. In particular, the clusters that arose in response to cuprizone in mice were similar to those in MS patients. Prinz noted, however, that other human microglial phenotypes might emerge if they expanded their analysis to MS patients in different stages or with varying types of disease.
“This study is very exciting,” wrote Jernej Ule, University College London, to Alzforum. “Some of the differences between mouse and human data could be technical, or due to differences in age of individuals, etc. However, the similarities are more striking to me,” he said.
Other investigators are pursing similar profiling studies, though consensus has yet to emerge about methodology and interpretation. In a paper currently online on bioRχiv, Philip De Jager’s group at Columbia University, New York reported 14 clusters among 15,910 microglia isolated from seven epilepsy surgical samples and eight samples of postmortem tissue from older adults, either non-demented or with Alzheimer’s, frontotemporal dementia, or Parkinson’s disease (Olah et al., 2018). They weren’t able to tie any cluster clearly to disease, perhaps due to the small number of patients. “The number of subjects providing such data remains limited, so it will be essential to assemble larger sample sets and compare data across centers to derive a robust model of the population structure of microglia,” wrote De Jager to Alzforum. He has made his group’s transcriptome data freely available in a searchable database.
Meanwhile other labs are taking alternative routes to characterize microglia diversity. One approach is single-nuclei RNA sequencing, which has the advantage of using frozen or fixed tissues (Jul 2018 conference news). Also promising—multiplexed antibody-based proteomics approaches that can identify dozens to hundreds of markers simultaneously on single cells in situ (Dec 2018 news).—Pat McCaffrey
References
News Citations
- In Pathology Cascade, Microglia Rev Up After Plaques but Before Tangles
- Microglia Reveal Formidable Complexity, Deep Culpability in AD
- A Delicate Frontier: Human Microglia Focus of Attention at Keystone
- Local Flavor: At Protein Level, Too, Human Microglia Are Diverse
Paper Citations
- Li Q, Cheng Z, Zhou L, Darmanis S, Neff NF, Okamoto J, Gulati G, Bennett ML, Sun LO, Clarke LE, Marschallinger J, Yu G, Quake SR, Wyss-Coray T, Barres BA. Developmental Heterogeneity of Microglia and Brain Myeloid Cells Revealed by Deep Single-Cell RNA Sequencing. Neuron. 2019 Jan 16;101(2):207-223.e10. Epub 2018 Dec 31 PubMed.
- Olah M, Menon V, Habib N, Taga M, Yung C, Cimpean M, Khairalla A, Dionne D, Hopp S, Frosch MP, Hyman BT, Beach TG, Sarkis R, Cosgrove GR, Helgager J, Golden JA, Pennell PB, Schneider JA, Bennett DA, Regev A, Elyaman W, Bradshaw EM, De Jager PL. A single cell-based atlas of human microglial states reveals associations with neurological disorders and histopathological features of the aging brain. bioRχiv. June 11, 2018 BioRxiv.
External Citations
Further Reading
No Available Further Reading
Primary Papers
- Masuda T, Sankowski R, Staszewski O, Böttcher C, Amann L, Sagar, Scheiwe C, Nessler S, Kunz P, van Loo G, Coenen VA, Reinacher PC, Michel A, Sure U, Gold R, Grün D, Priller J, Stadelmann C, Prinz M. Spatial and temporal heterogeneity of mouse and human microglia at single-cell resolution. Nature. 2019 Feb;566(7744):388-392. Epub 2019 Feb 13 PubMed.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.