New Stem Cell-Derived Neuron Datasets Available
Quick Links
It's time to add two new stem cell databases to the neurological disease research toolbox. For one, Clive Svendsen at Cedars-Sinai Medical Center, Los Angeles, and colleagues at Answer ALS led a team of scientists that created motor neurons from induced pluripotent stem cells derived from 92 healthy people and 341 with amyotrophic lateral sclerosis. In the February 9 Neuron, they reported disease signatures in the ALS neurons that were sex-dependent.
For the second, Cornelis Blauwendraat at the National Institute on Aging in Bethesda, Maryland, and colleagues induced dopaminergic neurons from iPSCs derived from 95 people, including some with sporadic PD, asymptomatic and symptomatic PD mutation carriers, and controls. The February 6 Cell Genomics contains a plethora of genetic, epigenetic, gene expression, and cell imaging data—the first set from the Foundational Data Initiative for Parkinson's Disease (FOUNDIN-PD). Co-corresponding authors on this international collaboration include Andrew Singleton and Mark Cookson at NIA, Peter Heutink and Vikas Bansal at Germany’s DZNE in Tübingen, David Craig at the University of Southern California in Los Angeles, Kendall Van Keuren-Jensen at the Translational Genomics Research Institute in Phoenix, and Steven Finkbeiner at the Gladstone Institutes in San Francisco.
A longitudinal study of 1,000 people with ALS and healthy controls, Answer ALS generates iPSCs from each participant's blood cells, then turns the stem cells into brain-like cells for genomic, transcriptomic, and proteomic analysis (see image below). Svendsen and co-corresponding authors Dhruv Sareen at Cedars-Sinai and Leslie Thompson of the University of California, Irvine, derived motor neurons from 433 of these volunteers. Thirty-seven of the 341 with ALS carried pathogenic variants in C9ORF72 or SOD1, while the rest had sporadic disease.
As described in Neuron, co-first authors Michael Workman, Ryan Lim, and Jie Wu found that stem cells from ALS participants generated more motor neurons than did control iPSCs, with the bulk of this difference coming from male cells. This sex difference intrigued the scientists, since ALS is more common in men, and their symptoms tend to begin earlier than in women.
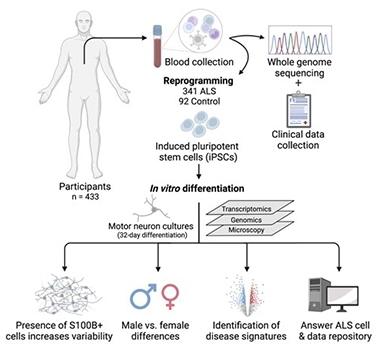
Answers From ALS Neurons? Blood cells from 433 donors were reprogrammed into iPSCs, then differentiated into motor neurons. The neurons were analyzed and the data made available via the Answer ALS repository online. [Courtesy of Workman et al., Neuron, 2023.]
This sex difference persisted in gene expression. While there were only a handful of differentially expressed genes in ALS versus control neurons in toto, 350 DEGs emerged when the scientists compared only motor neurons from men. Upregulated genes included those in the inflammatory TNF and NK-κB signaling pathways. There were no DEGs when comparing cells taken from women with ALS to those from healthy women. “The striking sex differences in our data suggest that males and females may need to be analyzed separately, requiring twice as many subjects for equivalently powered studies,” Workman and colleagues wrote.
The scientists also noticed that gene expression in the cultured neurons correlated with disease progression, as measured by the revised ALS Functional Rating Scale. For example, neurons expressing the least HSPBAP1, which is downregulated in the motor cortex of ALS cases, or NUP188, a target of TDP-43, came from people who had declined quickly (Saris et al., 2009; Roczniak-Ferguson et al., 2019). By measuring expression of those two genes, plus five others that tracked with ALSFRS score, the scientists predicted disease progression rate with 66 percent accuracy.
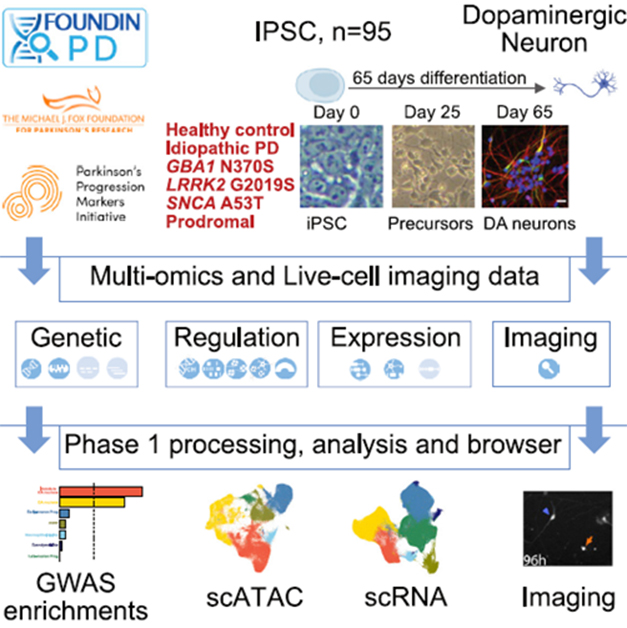
PD Neurons. Researchers differentiated iPSCs from healthy controls and people with PD into dopaminergic neurons, then analyzed the cells’ genomes, epigenomes, transcriptomes, and morphology. [Courtesy of Bressan et al., Cell Genomics, 2023.]
For the FOUNDIN-PD dataset, co-first authors Elisangela Bressan, Xylena Reed, and Vikas Bansal created dopaminergic neurons from iPSCs of nine healthy participants, 26 unaffected carriers of PD risk variants, four people with prodromal PD, and 56 people with PD enrolled in the Parkinson’s Progression Markers Initiative. PPMI is a longitudinal study following more than 1,400 people with and without PD in 11 countries. Of the unaffected carriers, 13 had a LRRK2 variant, 12 a GBA1 mutation, and one had an A53T SNCA mutation. PD participants included 13 LRRK2 carriers, eight GBA1, three SNCA, and 32 with idiopathic PD.
Bressan and colleagues sequenced the cells' genomes, ran single-cell RNA-Seq, measured DNA methylation, analyzed open chromatin with ATAC-seq, and captured morphological change and cell death over 10 days by microscopy (image above).
The transcriptomes of the dopaminergic neurons correlated strongly with those of tyrosine hydroxylase-positive neurons isolated postmortem from human substantia nigra, suggesting the cultured neurons recapitulate in vivo expression to some extent (see Agarwal et al., 2020). More neurons carrying a GBA1 mutation degenerated over eight days in culture than did wild-type control neurons. The scientists hope that further analysis of these cell lines will tease out how PD genetic variants disrupt molecular processes.
Access the ALS and PD repositories for further information.—Chelsea Weidman Burke
References
Paper Citations
- Saris CG, Horvath S, van Vught PW, van Es MA, Blauw HM, Fuller TF, Langfelder P, Deyoung J, Wokke JH, Veldink JH, van den Berg LH, Ophoff RA. Weighted gene co-expression network analysis of the peripheral blood from Amyotrophic Lateral Sclerosis patients. BMC Genomics. 2009;10:405. PubMed.
- Roczniak-Ferguson A, Ferguson SM. Pleiotropic requirements for human TDP-43 in the regulation of cell and organelle homeostasis. Life Sci Alliance. 2019 Oct;2(5) Print 2019 Oct PubMed.
- Agarwal D, Sandor C, Volpato V, Caffrey TM, Monzón-Sandoval J, Bowden R, Alegre-Abarrategui J, Wade-Martins R, Webber C. A single-cell atlas of the human substantia nigra reveals cell-specific pathways associated with neurological disorders. Nat Commun. 2020 Aug 21;11(1):4183. PubMed.
External Citations
Further Reading
Primary Papers
- Workman MJ, Lim RG, Wu J, Frank A, Ornelas L, Panther L, Galvez E, Perez D, Meepe I, Lei S, Valencia V, Gomez E, Liu C, Moran R, Pinedo L, Tsitkov S, Ho R, Kaye JA, Answer ALS Consortium, Thompson T, Rothstein JD, Finkbeiner S, Fraenkel E, Sareen D, Thompson LM, Svendsen CN. Large-scale differentiation of iPSC-derived motor neurons from ALS and control subjects. Neuron. 2023 Apr 19;111(8):1191-1204.e5. Epub 2023 Feb 9 PubMed.
- Bressan E, Reed X, Bansal V, Hutchins E, Cobb MM, Webb MG, Alsop E, Grenn FP, Illarionova A, Savytska N, Violich I, Broeer S, Fernandes N, Sivakumar R, Beilina A, Billingsley KJ, Berghausen J, Pantazis CB, Pitz V, Patel D, Daida K, Meechoovet B, Reiman R, Courtright-Lim A, Logemann A, Antone J, Barch M, Kitchen R, Yan Li Y, Dalgard CL, Rizzu P, Hernandez DG, Hjelm BE, Nalls M, Gibbs JR, Finkbeiner S, Cookson MR, Van Keuren-Jensen K, Craig DW, Singleton AB, Heutink P, Blauwendraat C. The Foundational Data Initiative for Parkinson Disease: Enabling efficient translation from genetic maps to mechanism. Cell Genomics, February 6, 2023
Annotate
To make an annotation you must Login or Register.

Comments
Harvard Medical School
Harvard Medical School
Studies of Mendelian inheritance have found genetic mutations that cause rare forms of PD, but iPSC-derived neural cells from members of such families are functional and require a strong “second challenge” to present cell biological phenotypes of PD, consistent with non-genetic factors, such as aging, conferring the strongest risk of disease (Polymeropoulos et al., 1997; Zimprich et al., 2004; Paisán-Ruíz et al., 2004; Valente et al., 2004; Cooper et al., 2012).
Toward extending our understanding of genomic influences for common idiopathic PD, GWAS have identified many nucleotide variations in blood that are associated with mild risk of PD (Nalls et al., 2019). Connecting these nucleotides to actionable genes and pathways in the aging human brain is a challenge.
Here Elisangela Bressan, Xylena Reed, Vikas Bansal, and the FOUNDIN-PD group (Bressan et al., 2023) generated 95 iPSCs from individuals enrolled in the Parkinson’s Progression Markers Initiative (PPMI), an invaluable cohort of volunteers whose symptoms are tracked over time. All iPSCs were differentiated into dopaminergic neurons using automated processes. High-dimensional biochemical analyses linked the rs11950533 nucleotide to CAMLG expression in dopaminergic neurons, showing an example of functional genomics during human developmental biology.
The addition of iPSCs from genetically diverse individuals to the initiative will be an important step toward building an inclusive public repository of cell lines that can be used to broaden our understanding of functional genomics during human developmental biology. However, the problem of linking genetic variation to gene expression in patient cell types remains largely unresolved. In fact, the emerging data for somatic mosaicism in the human brain may further challenge our ability to interpret polygenic risk scores derived from blood (Breuss et al., 2022; Pollina et al., 2023; Madabhushi et al., 2015; McKinnon, 2013).
High-confidence single-cell DNA sequencing of adult human brain regions that are vulnerable to degeneration will directly test any links between polygenic risk scores from blood and disease-relevant neurons. Dopaminergic neurons differentiated from iPSCs do not seem like a streamlined approach to understanding the effects of single nucleotide changes upon the function of adult dopaminergic neurons. Indeed, stochastic sequence variation during iPSC differentiation may dilute the already weak disease effects of a combined 200 or so nucleotides in a shifting genome consisting of 3 billion nucleotides (Nurk et al., 2022; Aganezov et al., 2022).
Investing resources to improve methods that confidently report the genomic sequences of single adult human neurons is a prudent technological step for PD research.
References:
Polymeropoulos MH, Lavedan C, Leroy E, Ide SE, Dehejia A, Dutra A, Pike B, Root H, Rubenstein J, Boyer R, Stenroos ES, Chandrasekharappa S, Athanassiadou A, Papapetropoulos T, Johnson WG, Lazzarini AM, Duvoisin RC, Di Iorio G, Golbe LI, Nussbaum RL. Mutation in the alpha-synuclein gene identified in families with Parkinson's disease. Science. 1997 Jun 27;276(5321):2045-7. PubMed.
Zimprich A, Biskup S, Leitner P, Lichtner P, Farrer M, Lincoln S, Kachergus J, Hulihan M, Uitti RJ, Calne DB, Stoessl AJ, Pfeiffer RF, Patenge N, Carbajal IC, Vieregge P, Asmus F, Müller-Myhsok B, Dickson DW, Meitinger T, Strom TM, Wszolek ZK, Gasser T. Mutations in LRRK2 cause autosomal-dominant parkinsonism with pleomorphic pathology. Neuron. 2004 Nov 18;44(4):601-7. PubMed.
Paisán-Ruíz C, Jain S, Evans EW, Gilks WP, Simón J, van der Brug M, López de Munain A, Aparicio S, Gil AM, Khan N, Johnson J, Martinez JR, Nicholl D, Carrera IM, Pena AS, de Silva R, Lees A, Martí-Massó JF, Pérez-Tur J, Wood NW, Singleton AB. Cloning of the gene containing mutations that cause PARK8-linked Parkinson's disease. Neuron. 2004 Nov 18;44(4) PubMed.
Valente EM, Abou-Sleiman PM, Caputo V, Muqit MM, Harvey K, Gispert S, Ali Z, Del Turco D, Bentivoglio AR, Healy DG, Albanese A, Nussbaum R, González-Maldonado R, Deller T, Salvi S, Cortelli P, Gilks WP, Latchman DS, Harvey RJ, Dallapiccola B, Auburger G, Wood NW. Hereditary early-onset Parkinson's disease caused by mutations in PINK1. Science. 2004 May 21;304(5674):1158-60. Epub 2004 Apr 15 PubMed.
Cooper O, Seo H, Andrabi S, Guardia-Laguarta C, Graziotto J, Sundberg M, McLean JR, Carrillo-Reid L, Xie Z, Osborn T, Hargus G, Deleidi M, Lawson T, Bogetofte H, Perez-Torres E, Clark L, Moskowitz C, Mazzulli J, Chen L, Volpicelli-Daley L, Romero N, Jiang H, Uitti RJ, Huang Z, Opala G, Scarffe LA, Dawson VL, Klein C, Feng J, Ross OA, Trojanowski JQ, Lee VM, Marder K, Surmeier DJ, Wszolek ZK, Przedborski S, Krainc D, Dawson TM, Isacson O. Pharmacological rescue of mitochondrial deficits in iPSC-derived neural cells from patients with familial Parkinson's disease. Sci Transl Med. 2012 Jul 4;4(141):141ra90. PubMed.
Nalls MA, Blauwendraat C, Vallerga CL, Heilbron K, Bandres-Ciga S, Chang D, Tan M, Kia DA, Noyce AJ, Xue A, Bras J, Young E, von Coelln R, Simón-Sánchez J, Schulte C, Sharma M, Krohn L, Pihlstrøm L, Siitonen A, Iwaki H, Leonard H, Faghri F, Gibbs JR, Hernandez DG, Scholz SW, Botia JA, Martinez M, Corvol JC, Lesage S, Jankovic J, Shulman LM, Sutherland M, Tienari P, Majamaa K, Toft M, Andreassen OA, Bangale T, Brice A, Yang J, Gan-Or Z, Gasser T, Heutink P, Shulman JM, Wood NW, Hinds DA, Hardy JA, Morris HR, Gratten J, Visscher PM, Graham RR, Singleton AB, 23andMe Research Team, System Genomics of Parkinson's Disease Consortium, International Parkinson's Disease Genomics Consortium. Identification of novel risk loci, causal insights, and heritable risk for Parkinson's disease: a meta-analysis of genome-wide association studies. Lancet Neurol. 2019 Dec;18(12):1091-1102. PubMed.
Bressan E, Reed X, Bansal V, Hutchins E, Cobb MM, Webb MG, Alsop E, Grenn FP, Illarionova A, Savytska N, Violich I, Broeer S, Fernandes N, Sivakumar R, Beilina A, Billingsley KJ, Berghausen J, Pantazis CB, Pitz V, Patel D, Daida K, Meechoovet B, Reiman R, Courtright-Lim A, Logemann A, Antone J, Barch M, Kitchen R, Li Y, Dalgard CL, The American Genome Center, Rizzu P, Hernandez DG, Hjelm BE, Nalls M, Gibbs JR, Finkbeiner S, Cookson MR Van Keuren-Jensen K, Craig DW Singleton AB, Heutink P, Blauwendraat C. The Foundational Data Initiative for Parkinson Disease: Enabling efficient translation from genetic maps to mechanism. Cell Genomics 3, March 8, 2023 Cell Genomics
Breuss MW, Yang X, Schlachetzki JC, Antaki D, Lana AJ, Xu X, Chung C, Chai G, Stanley V, Song Q, Newmeyer TF, Nguyen A, O'Brien S, Hoeksema MA, Cao B, Nott A, McEvoy-Venneri J, Pasillas MP, Barton ST, Copeland BR, Nahas S, Van Der Kraan L, Ding Y, NIMH Brain Somatic Mosaicism Network, Glass CK, Gleeson JG. Somatic mosaicism reveals clonal distributions of neocortical development. Nature. 2022 Apr;604(7907):689-696. Epub 2022 Apr 20 PubMed.
Pollina EA, Gilliam DT, Landau AT, Lin C, Pajarillo N, Davis CP, Harmin DA, Yap EL, Vogel IR, Griffith EC, Nagy MA, Ling E, Duffy EE, Sabatini BL, Weitz CJ, Greenberg ME. A NPAS4-NuA4 complex couples synaptic activity to DNA repair. Nature. 2023 Feb;614(7949):732-741. Epub 2023 Feb 15 PubMed.
Madabhushi R, Gao F, Pfenning AR, Pan L, Yamakawa S, Seo J, Rueda R, Phan TX, Yamakawa H, Pao PC, Stott RT, Gjoneska E, Nott A, Cho S, Kellis M, Tsai LH. Activity-Induced DNA Breaks Govern the Expression of Neuronal Early-Response Genes. Cell. 2015 Jun 18;161(7):1592-605. Epub 2015 Jun 4 PubMed.
McKinnon PJ. Maintaining genome stability in the nervous system. Nat Neurosci. 2013 Nov;16(11):1523-9. Epub 2013 Oct 28 PubMed.
Nurk S, Koren S, Rhie A, Rautiainen M, Bzikadze AV, Mikheenko A, Vollger MR, Altemose N, Uralsky L, Gershman A, Aganezov S, Hoyt SJ, Diekhans M, Logsdon GA, Alonge M, Antonarakis SE, Borchers M, Bouffard GG, Brooks SY, Caldas GV, Chen NC, Cheng H, Chin CS, Chow W, de Lima LG, Dishuck PC, Durbin R, Dvorkina T, Fiddes IT, Formenti G, Fulton RS, Fungtammasan A, Garrison E, Grady PG, Graves-Lindsay TA, Hall IM, Hansen NF, Hartley GA, Haukness M, Howe K, Hunkapiller MW, Jain C, Jain M, Jarvis ED, Kerpedjiev P, Kirsche M, Kolmogorov M, Korlach J, Kremitzki M, Li H, Maduro VV, Marschall T, McCartney AM, McDaniel J, Miller DE, Mullikin JC, Myers EW, Olson ND, Paten B, Peluso P, Pevzner PA, Porubsky D, Potapova T, Rogaev EI, Rosenfeld JA, Salzberg SL, Schneider VA, Sedlazeck FJ, Shafin K, Shew CJ, Shumate A, Sims Y, Smit AF, Soto DC, Sović I, Storer JM, Streets A, Sullivan BA, Thibaud-Nissen F, Torrance J, Wagner J, Walenz BP, Wenger A, Wood JM, Xiao C, Yan SM, Young AC, Zarate S, Surti U, McCoy RC, Dennis MY, Alexandrov IA, Gerton JL, O'Neill RJ, Timp W, Zook JM, Schatz MC, Eichler EE, Miga KH, Phillippy AM. The complete sequence of a human genome. Science. 2022 Apr;376(6588):44-53. Epub 2022 Mar 31 PubMed.
Aganezov S, Yan SM, Soto DC, Kirsche M, Zarate S, Avdeyev P, Taylor DJ, Shafin K, Shumate A, Xiao C, Wagner J, McDaniel J, Olson ND, Sauria ME, Vollger MR, Rhie A, Meredith M, Martin S, Lee J, Koren S, Rosenfeld JA, Paten B, Layer R, Chin CS, Sedlazeck FJ, Hansen NF, Miller DE, Phillippy AM, Miga KH, McCoy RC, Dennis MY, Zook JM, Schatz MC. A complete reference genome improves analysis of human genetic variation. Science. 2022 Apr;376(6588):eabl3533. PubMed.
University of Miami Miller School of Medicine
In this study, Bressan and colleagues took an iPSC approach to better understand the impact genetic factors, whether resulting from single-gene variants or a combination of PD-associated variants, play in PD pathobiology. Specifically, it sought to understand how genetic risk factors—many of which may be noncoding and with unknown effects on disease—may alter the function and dysfunction of dopaminergic neurons. This first data release by the Foundational Data Initiative for Parkinson Disease (FOUNDIN-PD) collaborative serves as a blueprint for the approach this team has developed to tease apart the complexity of PD biology. It uses multiple, genetically defined iPSC lines from affected and unaffected individuals differentiated into dopaminergic neurons using a consistent, validated method followed by implementation of an extensive battery of high-throughput phenotypic characterization methods to generate complementary data sets of epigenetic, regulatory, transcriptional, and cellular imaging data. This data was subjected to robust, integrated data analysis approaches and made available to the broader research community through the FOUNDIN-PD data portal.
Many studies have used iPSC-derived DA neurons to study the impact of specific genetic variants on PD pathobiology. Results have often been difficult to compare due to differences in the genetic background of the iPSC lines, differentiation approaches used to generate the DA neurons, and phenotypes being analyzed, as well as the small sample sizes available.
This study used strategies to generate more useful and applicable data and to minimize the heterogeneity implicit in studies of human samples. Samples were drawn from PPMI, hence had been extensively characterized. In this first analysis, 95 iPSC lines were used for differentiation and characterization. This included individuals bearing disease-associated monogenic variants in LRRK2 (26; 13 with PD, 13 carriers), GBA1 (20; eight with PD, 12 carriers), and SNCA (four; three with PD, one carrier). Individuals with idiopathic PD (32) were chosen based on whether they had a high or low polygenic risk score (PRS). Additionally, iPSC lines from nine healthy, unaffected and four prodromal/SWEDD (scans without evidence for dopaminergic deficits) individuals were included.
To minimize heterogeneity associated with DA differentiation, the authors used an optimized protocol that could be carried out on an automated robotic cell culture system. That allowed for multiple iPSC lines to be differentiated at the same time, minimizing batch-to-batch variability in the efficacy of DA neuron production. A pilot validation study of this protocol produced >60 percent tyrosine hydroxylase (TH)-positive neurons; TH is a marker for DA neurons. This was significantly lower (~20 percent TH+ neurons) when the PPMI iPSC lines underwent differentiation. However, by employing high-throughput approaches for the functional analyses, including single-cell, they were able to generate robust data across multiple parameters despite this low yield.
In this initial study, genetic analysis generated genotype data and mitochondrial DNA sequencing; the cells' regulatory architecture was analyzed for DNA methylation patterns, chromatin accessibility (ATACseq and single-cell ATACseq) and chromatin conformation mapping (Hi-C), which could be compared to transcriptional outcomes (small RNA-Seq, RNA-Seq, and single-cell RNA-Seq). The combination of these analyses facilitated identification of expression quantitative trait loci (eQTL) that may help to narrow down putative genetic drivers of PD from genome-wide association study (GWAS) loci. Finally, longitudinal imaging analysis using robotic microscopy was performed to track changes in individual neuronal features, with the main analysis focusing on neuronal survival.
This paper focused on describing the optimization of the approach FOUNDIN-PD will use to dissect cellular and molecular mechanisms that underlie PD through standardized derivation of DA neurons from large numbers of iPSC lines followed by phenotypic and integrated data analyses approaches, providing datasets that can serve the research community and accelerate discoveries toward therapeutic approaches. As this group moves forward, I expect their approaches will mature and increase in scale to produce exceedingly more comprehensive and well-validated datasets.
As the authors suggest, additional cell types that contribute to PD pathology could be examined with similar approaches. It would be nice to see this group incorporate genome-editing approaches into the workflow to produce isogenic pairs of iPSC lines that differ only in the variant(s) of interest and not in effects driven by differences in the genetic architecture between individuals. This would be most easily achieved for monogenic forms of PD but could be done based on genes/variants that are contributing to the PRS.
Incorporation of iPSC lines derived from individuals of different ancestral backgrounds would broaden the reach, provide complementary data that may reveal new pathways or strengthen the association with other common pathways, and maximize benefits to the PD population garnered from these studies. Altogether, this manuscript provides a roadmap for exploring molecular drivers of PD with the intention of identifying therapeutic targets that will correct the cellular defects that underlie PD rather than simply treat pathological and clinical outcomes of the disease process.
University of Cambridge
PD and other neurodegenerative diseases have been studied for decades but treatments that effectively target disease mechanisms remain few and far between. It is therefore greatly encouraging to see collaborative initiatives like FOUNDIN-PD that enable ambitious, large-scale, team science approaches that are mapping out the perturbations seen in disease in unprecedented detail and promise to reveal new therapeutic targets. Specifically, convergence of GWAS-implicated genes onto pathways highlighted by familial mutations is fertile ground for new biological discovery. The open-science approach of making data readily available via a browser will certainly be of use to other groups in this field.
The authors’ use of standardized and well-established directed differentiation methods and robotic liquid handling limited line-to-line variability in differentiation outcomes. However, as in other studies, this variability did persist. It may complicate the interpretation of bulk RNA-Seq and ATAC-Seq data, so the use of single-cell approaches is a great idea. The scale of the resulting single-cell dataset (more than 400,000 cells) enabled the identification of candidate genetic variants that might play cell-type-specific roles relevant to PD via MAGMA and an eQTL mapping. This revealed signals enriched in dopaminergic neurons at candidate risk genes, including CAMLG, TBC1D5, CCAR2, and ARIH2. Thus, though the consortium is still in its early stages, biological insights are already being made, and the scope and scale of analysis modalities extend well beyond earlier efforts using similar approaches (Jerber et al., 2021).
Looking ahead, it would be ideal to replace the long duration and cellular heterogeneity that results from directed dopaminergic differentiation protocols with a transcription-factor-based forward programming approach. While these methods have not yet reached maturity for dopaminergic neurons, the cellular resource and collaborative team of FOUNDIN-PD are well-placed to exploit them, and it would be interesting to see how results compare across differentiation methods.
References:
Jerber J, Seaton DD, Cuomo AS, Kumasaka N, Haldane J, Steer J, Patel M, Pearce D, Andersson M, Bonder MJ, Mountjoy E, Ghoussaini M, Lancaster MA, HipSci Consortium, Marioni JC, Merkle FT, Gaffney DJ, Stegle O. Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat Genet. 2021 Mar;53(3):304-312. Epub 2021 Mar 4 PubMed.
View all comments by Florian MerkleUniversity of Cambridge
It’s exciting to see this resource paper from members of the collaborative AnswerALS consortium, which has the ambitious goal to generate and utilize a biorepository of iSPCs from individuals suffering from ALS and unrelated controls to gain insight into disease processes via deep genotypic and phenotypic characterization. In this study, hundreds of iPSCs were differentiated toward motor neurons, the cell types most affected in ALS, resulting in cultures containing a mixture of motor neuron-like cells and other cell types.
Other consortia such as iNDI (Ramos et al., 2021) are engineering some ALS-associated mutations onto the same genetic background. It is very valuable to have a complementary resource, such as the one described in this paper, to provide insight into how similar mutations may have variable effects on different genetic backgrounds, and to identify commonly perturbed pathways that might emerge. While it would have been encouraging to see a stronger effect of genotype on cellular phenotypes, the promise of further analysis of the same samples across distinct modalities offers an exciting opportunity to gain insight into disease mechanisms.
One key finding is the split between male and female iPSC-derived spinal cord cultures in principal component analysis, which is remarkable in its consistency and effect size. The magnitude of this effect was significantly reduced when only autosomal genes were examined; even so, the finding is likely real since it was confirmed by analyzing primary human brain samples and could not be fully explained by erosion of X chromosome inactivation. This finding argues strongly for inclusion of cell lines derived from both sexes in future studies, or at least replication of key findings made in a cell line of a certain sex in another cell line of the other sex.
A major outstanding question is the extent to which changes in the differentiation protocol might alter the outcomes reported in this study. The authors clearly took great pains to consult the field, used well-established methods, and included batch differentiation controls to account for potential sources of variability. However, there are fundamental limitations to directed differentiation. First, since iPSC clones can have distinct intrinsic neuronal differentiation propensities, sometimes even among sister clones derived from the same donor cell pool that are nearly genetically identical (Jerber et al., 2021), it can be challenging to confidently assign changes in differentiation efficiency or transcriptional state to donor genotype.
Second, even when great effort is made to harmonize protocols or carry out an experiment under controlled conditions, as was done in this instance, the outcome of differentiation protocols can vary between users and institutions. While the current study was carried out with stringent controls in mind, one does wonder how reproducible some of the key findings might be if differentiations were performed again elsewhere.
Therefore, it would be of great interest to replicate these findings using methods such as inducible-transcription-factor-based forward programming, using methods such as piggyBac-based transgenesis, which allow for efficient and stable integration of reprogramming cassettes. While these methods have their own weaknesses, the resulting cell types are often more homogeneous and consistent across differentiation batches, enabling the effects of genotype to be compared across similar cell types, which hopefully would allow some of the residual variation reported in the current study (76.4 percent) to be assigned to genotype and other categories that give more mechanistic insight into ALS disease biology.
References:
Ramos DM, Skarnes WC, Singleton AB, Cookson MR, Ward ME. Tackling neurodegenerative diseases with genomic engineering: A new stem cell initiative from the NIH. Neuron. 2021 Apr 7;109(7):1080-1083. PubMed.
Jerber J, Seaton DD, Cuomo AS, Kumasaka N, Haldane J, Steer J, Patel M, Pearce D, Andersson M, Bonder MJ, Mountjoy E, Ghoussaini M, Lancaster MA, HipSci Consortium, Marioni JC, Merkle FT, Gaffney DJ, Stegle O. Population-scale single-cell RNA-seq profiling across dopaminergic neuron differentiation. Nat Genet. 2021 Mar;53(3):304-312. Epub 2021 Mar 4 PubMed.
View all comments by Florian MerkleMake a Comment
To make a comment you must login or register.