First Whole-Genome Sequencing of PSP Nets Six New Risk Loci
Quick Links
Progressive supranuclear palsy, a rare tauopathy, slowly destroys a person's cognition, balance, and movement. PSP ranks second behind Parkinson’s disease as a cause of Parkinsonism. Beyond variants in the tau gene, a handful of other risk genes have turned up in about 10 genome-wide association studies. Now, scientists led by Wan-Ping Lee and Gerard Schellenberg of the University of Pennsylvania in Philadelphia; Daniel Geschwind at the University of California, Los Angeles; Günter Höglinger of the Ludwig-Maximilians University Hospital in Munich; and Dennis Dickson at Mayo Clinic, Jacksonville, Florida, report the first whole-genome sequencing of PSP in a preprint uploaded to medRxiv on January 30.
- Scientists sequenced the genomes of 1,700 people with PSP.
- They found six known and six new risk loci.
- APOE2 seemed to increase risk.
They uncovered new common, rare, and structural variant among 12 loci, including APOE. Curiously, APOE2 seemed to up a person’s odds of PSP, in contrast to its protective role in Alzheimer’s disease.
James Rowe of the University of Cambridge, U.K., thinks this study is important. “These discoveries can now be mapped to pathogenic pathways to guide much-needed therapeutic strategies,” he wrote to Alzforum (comment below).
While previous GWAS uncovered common risk variants, GWAS generally fall short of finding rare mutations, including small insertions and deletions (indels), and larger structural variants (Jun 2011 news; Nov 2023 news). To get a sense of how such variants influence PSP, co-first authors Hui Wang of UPenn and Timothy Chang at UCLA analyzed whole-genome sequencing data of 1,718 people with PSP and 2,944 controls from the National Institute on Aging’s Alzheimer’s Disease Sequencing Project, a collection of genetic data from 40 cohorts worldwide. On average, people with PSP were in their mid-60s, while controls were in their 80s. All participants were of European ancestry and half were women. PSP was autopsy-confirmed in 1,441 cases and clinically diagnosed in the rest.
The WGS turned up six loci previously linked to PSP, including the tau gene. MAPT harbored common single-nucleotide variants (SNVs), indels, and a 238-bp deletion within intron 9. MAPT was the strongest risk gene for PSP in this and previous studies (image below). Common SNVs and indels in the myelin-associated protein gene MOBP and in STX6, which encodes a SNARE protein involved in intracellular trafficking, also reached genome-wide significance, i.e., a p-value of less than 5 x 10-8.
Falling short with p-values below 1 x 10-6, which the authors deemed "suggestive of significance,” were the phosphatase gene DUSP10, the transcription factor gene SP1, and the ion transporter gene SLCO1A2.
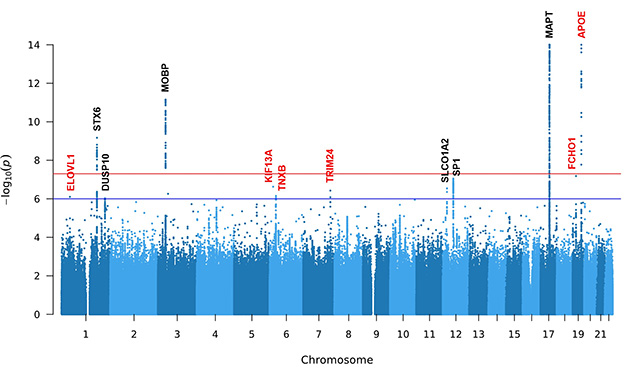
Genetic Skyline. This Manhattan plot shows common single-nucleotide variants and indels in six known loci (black) and six new ones (red). Only three known genes—MAPT, MOBP, and STX6—and the new loci APOE reached genome-wide significance (red horizontal line). The rest were suggestive (blue horizontal line). [Courtesy of Wang et al., medRxiv, 2024.]
Wang and Chang also found common SNVs and indels in six novel loci: APOE, the lipid elongase gene ELVOL1, the immunity gene FCHO1, the microtubule motor protein gene KIF13A, the extracellular matrix glycoprotein gene TNXB, and the transcription factor gene TRIM24 (image above). Six structural variants not previously linked to PSP occurred in five other loci. The most notable was a 619-bp deletion within PCMT1, which encodes a protein carboxyl methyltransferase highly expressed in the brain. Having the deletion in one copy of PCMT1 upped a person’s odds of having PSP by 4.2-fold; two copies increased the risk by 8.4 times.
The other four loci comprised the sperm protein gene SMCP, the histone acetyltransferase complex gene KANSL1, the immunoglobulin heavy gene, and a 1.5-kb stretch between the cytochrome P450 genes CYP2F1 and CYP2A13. KANSL1 was previously linked to tauopathies (Nov 2018 conference news; Mar 2022 conference news). As for rare variants, missense or splice mutations turned up in the zinc finger protein gene ZNF592. It has not been associated with PSP but controls the transcription of genes within the cerebellum, an area of the brain that regulates movement and balance (NIH gene entry).
ELVOL1 produces neurotoxic lipids in astrocytes, the cells that crank out most of the brain's APOE (Oct 2021 news). This led Lance Johnson and Lesley Golden of the University of Kentucky, Lexington, to wonder if the two PSP risk alleles conspire. “Does this suggest a common astrocyte lipoprotein-based mechanism of neurodegeneration in PSP?” they asked (comment below).
Most striking was the direction of APOE’s relationship to PSP: APOE2 seemed to make it go up. People with PSP were four times as likely to carry two copies of APOE2 than controls. Guojun Bu of Hong Kong University of Science and Technology, China, found the same in a cohort of 2,500 people with PSP. APOE2 carriers with PSP also had more tau pathology postmortem than did PSP cases with other APOE alleles. Bu thinks two cysteine residues within ApoE2 form disulfide bonds with two cysteines in tau (Zhao et al., 2018). This might shift the shape of tau’s fibrils, possibly helping explain the distinct cryo-EM structures of tau seen in PSP versus AD (Oct 2021 news).
While APOE4 seemed protective against PSP—again the opposite of AD—this did not hold when Lee excluded from his analysis cohorts with a higher prevalence of APOE4 carriers than in the general population. Because some of the cohorts' control groups still had unusual APOE allele frequencies after excluding outliers, Yadong Huang, University of California, San Francisco, thinks the direction of the APOE risk data needs to be confirmed in large cohorts.
Indeed, David Holtzman, Washington University, St. Louis, suspects the APOE associations might reflect selection bias. Because APOE4 increases the risk of amyloid deposition starting in mid-life and APOE2 reduces it, many people carrying APOE4 will go on to develop AD pathology, while APOE2 carriers will not. “I wonder then, if the pool of individuals who might get PSP is relatively enriched for APOE2,” he wrote (comment below).—Chelsea Weidman Burke
References
Mutations Citations
News Citations
- GWAS Fingers Tau and Other Genes for Parkinsonian Tauopathy
- In Progressive Supranuclear Palsy, Risk Loci Converge on Oligodendrocytes
- International Symposium Puts PSP/CBD on the Map
- Tau Haplotypes Hint at Transcriptional Changes, Ferroptosis
- ELOVL Hurts—Enzyme Makes Lipids That Turn Astrocytes Toxic
- Flock of New Folds Fills in Tauopathy Family Tree
Paper Citations
- Zhao N, Liu CC, Van Ingelgom AJ, Linares C, Kurti A, Knight JA, Heckman MG, Diehl NN, Shinohara M, Martens YA, Attrebi ON, Petrucelli L, Fryer JD, Wszolek ZK, Graff-Radford NR, Caselli RJ, Sanchez-Contreras MY, Rademakers R, Murray ME, Koga S, Dickson DW, Ross OA, Bu G. APOE ε2 is associated with increased tau pathology in primary tauopathy. Nat Commun. 2018 Oct 22;9(1):4388. PubMed.
External Citations
Further Reading
Primary Papers
- Wang H, Chang TS, Dombroski BA, Cheng P-L, Patil V, Valiente-Banuet L, Farrell K, McLean C, Molina-Porcel L, Rajput A, PaulDeDeyn P, LeBastard N, Gearing M, DonkerKaat L, VanSwieten JC, Dopper E, Ghetti BF, Newell KL, Troakes C, GdeYebenes J, Rabano-Gutierrez A, Meller T, Oertel WH, Respondek G, Stamelou M, Arzberger T, Roeber S, Muller U, Hopfner F, Hopfner F, Pastor P, Brice A, Durr A, LeBer I, Beach TG, Serrano GE, Hazrati L-N, Litvan I, Rademakers R, Ross OA, Galasko D, Boxer AL. Whole-Genome Sequencing Analysis Reveals New Susceptibility Loci and Structural Variants Associated with Progressive Supranuclear Palsy. 2024 Jan 30 10.1101/2023.12.28.23300612 (version 2) medRxiv.
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.