Artificial Intelligence Is Everywhere You Look in Dementia Research
Quick Links
Here’s a question on every Alzheimer’s disease researcher’s mind: How is artificial intelligence being used in dementia research? The prompt answer: "Artificial intelligence (AI) is playing a significant role in dementia research, helping scientists and medical professionals understand the disease better, improve diagnosis, and develop more effective treatments."
- AI tools can sift through large, multidimensional datasets to uncover hidden patterns.
- ChatGPT and other language processing AIs can detect hints of dementia in speech.
- AI is being used to analyze brain scans, neuropathology, protein folding, and to model AD pathogenesis.
Thanks ChatGPT, we’ll take it from here.
As AI programs like ChatGPT rise into mainstream consciousness, all but the stubbornest Luddites are starting to grasp how powerful, and often helpful, this technology can and will be. Beyond shallow or nefarious uses, like whipping up fake research reports on the fly, AI is already being harnessed in myriad aspects of scientific research, and the Alzheimer’s and dementia field is no exception. Scientists are using AI to merge multiomics datasets and plumb their depths. AI helps scientists uncover disease mechanisms and drug targets, see hidden features lurking among thousands of MRI scans, and understand what shapes amyloids into their fibrillar folds. AI is also proving useful in biomarker discovery, and detecting hints of impending dementia in speech before humans notice anything is amiss.
Reflecting AI’s push into all aspects of research and society, the European Parliament recently voted to move forward with the AI Act, a law that aims to safeguard human rights and ensure ethical development of AI in Europe. “The vote echoes a growing chorus of researchers from various disciplines calling for guardrails to govern powerful AI,” wrote Urs Gasser of the Technical University of Munich, in a recent perspective in Science (Gasser, 2023). Meanwhile, in the U.S., a recent study found that about 12 percent of approved medical devices that claim to use AI technologies had not been properly described as AI-enabled devices in their application to the FDA, suggesting the need for clear regulatory guidelines for this technology (Clark et al., 2023; Shah and Mello, 2023).
Despite its current celebrity status brought on by ChatGPT, AI itself is nothing new. In fact, scientists have been steadily improving upon AI methods such as machine learning, deep neural networks, and large language networks for decades. Equipped with layers of “neurons” that store and learn from different types of inputs, deep-learning techniques allow computers to process, learn, and extract hidden patterns from reams of complex, multidimensional data.
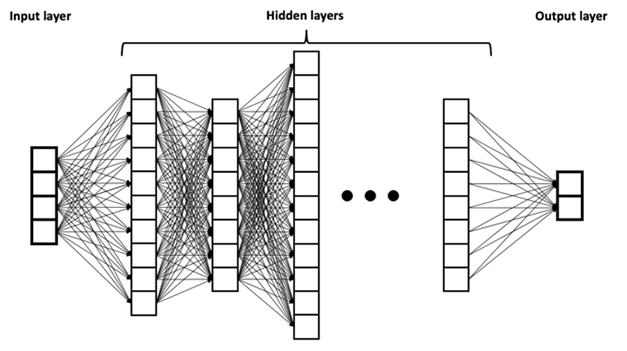
Layers of Learning. An example of a deep neural network, a type of AI method in which input data is processed by layers of “neurons” that store, process, and learn from the information, ultimately leading to an output for the user. [Courtesy of BruneloN, Wikimedia Commons.]
Yet AI has surged in the past decade. Why? According to Roland Eils, who leads the artificial intelligence working group at Berlin's Institute of Health at Charité-Universitätsmedizin, burgeoning amounts of interconnected, accessible data produced around the globe, plus recent advances in computational power and processing speed, culminated in the current boom. “These two factors together have led to a rebirth and explosion of the AI field,” Eils told Alzforum. “The amount of data is decisive.”
For example, Sjors Scheres of the MRC Laboratory of Molecular Biology, Cambridge, England, U.K., recently developed a machine-learning program, called ModelAngelo, to model the atomic structures of amyloidogenic proteins. “Machine learning is a rapidly developing field, with ever more powerful algorithms and network designs appearing constantly,” Scheres said.
Riding the Omics Tsunami
The neurodegenerative disease field is a prime example of this data surge, in terms of both its quantity and depth. “Deploying AI methods is just beginning in the field of dementia research, in which we have a rapidly growing ecosystem of multidimensional data that is becoming increasingly complex,” wrote Philip De Jager of Columbia University in New York.
To De Jager and others who study omics—a little suffix preceded by a growing lexicon—big, complex datasets are not merely a problem to deal with, but a necessary ingredient to make meaningful discoveries. To beef up sample size for different analyses requires harmonization of data collected from different cohorts, a necessity that poses Herculean statistical challenges. This is one place where AI comes in handy, De Jager said. “AI can have a very useful role in stitching together such datasets to create a larger, shared framework that can then be explored,” he wrote.
Case in point: De Jager's recent study used AI to look beneath cohort-level differences for links between gene expression and human phenotypes. Led by Su-In Lee and Sara Mostafavi at the University of Washington in Seattle, the study deployed a framework called Multi-task Deep learning for AD neuropathology (MD-AD), and identified sex-specific relationships between microglial immune responses and neuropathology (Beebe-Wang et al., 2021).
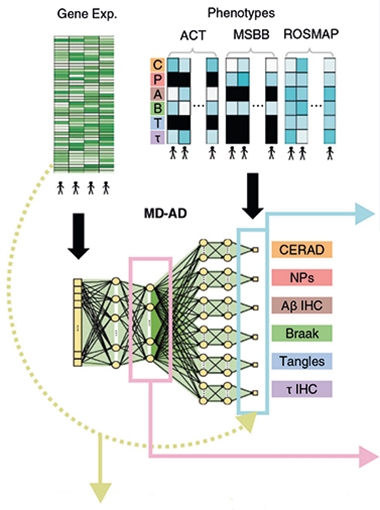
Combining Cohorts. Scientists fed gene expression and neuropathology data from participants in three cohorts into the MD-AD deep learning algorithm, which extracted a connection between expression and pathology features. [Courtesy of Beebe-Wang et al., Nature Communications, 2021.]
Thanks to the steadily growing sample size of GWAS, the number of risk variants for AD and other diseases has risen over the past decade (Feb 2021 news; Sep 2021 news). For these variant lists to be of use in discovering disease mechanisms and therapeutic targets, scientists need to understand their functional consequences. How does each risk variant influence expression and/or function of genes? In which cell types are they expressed? How does that promote or prevent disease? This is a tall order, given that the majority of risk variants reside within noncoding stretches of the genome. Deciphering how each signal does its work is a prime use for AI (for review, see Long et al., 2023). For example, one deep-learning model, called Sei, was trained to predict more than 20,000 regulatory features, such as transcription factor binding and chromatin accessibility, from more than 1,300 cell lines and tissue datasets (Chen et al., 2022). By scanning the entire genome for such features, Sei was then able to predict the effect of genetic variants in those regions.
AI is proving useful in generating more accurate polygenic risk scores. PRS are typically calculated by tallying the smidgeons of disease risk imparted by all genetic variants a person carries (Escott-Price et al., 2015; Jul 2016 news; Mar 2017 news). Thus far, PRS do not account for how variants interact with each other to influence risk, but a recent study used machine learning to factor in such epistatic effects. Researchers led by Nancy Ip of Hong Kong University of Science and Technology reported that a deep neural network model—trained on 8,100 SNP genotypes from more than 11,000 people in three cohorts—outperformed traditional statistical methods in predicting AD risk. What’s more, by sorting the contributing SNPs into biological categories and correlating them with levels of plasma proteins, the neural network helped pinpoint distinct mechanisms underlying disease risk, such as inflammation (Zhou et al., 2023).
What's next with this? “We envision that with further validation and optimization in larger cohorts, our model could serve as a screening tool in routine clinical prognosis and diagnosis, improving the accuracy and confidence in AD diagnosis,” Ip commented. “In addition, these tools may enable evaluation of drug response in people with different genetic risk levels.”
Others are deploying AI to predict drug targets from large public omics datasets. Take the Open Targets Platform. This public-private partnership integrates mind-boggling amounts of GWAS, single-cell sequencing, tissue expression, and functional genomics data to link GWAS hits to target genes, zero in on disease mechanisms, and predict and prioritize drug targets (Han et al., 2022). InSilico Medicine, a Hong Kong-based company that wields AI for drug discovery, recently used its algorithm, called PandaOmics, to predict 17 drug targets for ALS, including 11 novel ones (Pun et al., 2022).
Besides genetics, other factors, including many that are modifiable, also influence disease risk. AI is particularly handy at merging genetics with other types of health data to create a clearer picture. This type of multidimensional analysis is becoming possible thanks to large observational cohorts. With its half-million participants, the U.K. Biobank is a prime example. Volunteers undergo comprehensive health and cognitive exams, offer up blood and urine for cellular, genetic, and metabolite analysis, and lie in the scanner for multiorgan MRI. The resulting flood of data is amenable to machine learning.
Unleashing a deep neural network model on the UKBB data, the Charité's Eils and colleagues integrated cardiac polygenic risk assessment with clinical predictors of cardiac stress to foretell a person’s risk of heart attack within the following decade (Steinfeldt et al., 2022). Called NeuralCVD, it outperformed standard approaches, such as Cox hazard ratios, in predicting impending cardiac events. Eils’ group also applied machine learning to tie plasma metabolites to the future onset of 23 different diseases, including dementia and cognitive decline (Oct 2022 news).
Machine learning algorithms detected clinically diagnosed and prodromal Parkinson’s disease among 100,000 U.K. Biobank participants who agreed to have their every move tracked with an accelerometer for one week. This digital biomarker outperformed measures of genetics, lifestyle, or blood biochemistry up to seven years prior to diagnosis (Schalkamp et al., 2023).
Other scientists have plumbed UKBB's multi-organ imaging data. One recent study used AI to scour relationships between brain and heart MRI scans from 40,000 people, and correlate these with genetic variants (Jun 2023 news). The study identified thousands of connections between the two organs, as well as common genetic ties to disease. However, it also laid bare a problem in interpretating AI-driven findings: What causes what often remains unclear. While machine learning uncovered myriad connections between heart and brain, the task of discerning which of those are meaningful still falls to scientists.
An example of a more immediately practical application of AI is unfolding at University College London, where scientists led by Nick Fox are using machine learning to help them recognize patterns generated by new, more powerful forms of MRI. Their goal is to shorten the time a person spends in the scanner to well under a minute. This will boost MRI capacity as anti-amyloid antibody therapy is being rolled out.
Betty Tijms of Amsterdam University Medical Center uses fluid proteomics data to discern different AD subtypes and discover biomarkers. Tijms can't imagine doing her research without AI (Tijms et al., 2023; Gogishvili et al., 2023). Still, she noted that while AD makes data analysis efficient and less noisy, “biological intelligence” comes first in discovering new disease mechanisms. In other words, a person. AI makes it easier to model many variables at once, which helps generate hypotheses that must be put to the test at the bench.

Neuropath Mapping. A machine-learning algorithm learned to spot dense and diffuse Ab plaques, as well as neurofibrillary tangles, which could be mapped and quantified in human brain sections. [Courtesy of Stephen et al., BioRxiv, 2023.]
In some areas of research, the efficiency, consistency, and objectivity of machine intelligence is a respite from the slow and subjective inspections done by people. Neuropathology is an example. Researchers led by Michael Bienkowski at the University of Southern California in Los Angeles recently trained a machine-learning model to hunt down and map the distribution of Aβ plaques and tau tangles in hippocampal brain sections (Stephen et al., 2023). The algorithm distinguished between dense and diffuse Aβ plaques. “Diffuse things, by their nature, are difficult to count,” Bienkowski told Alzforum. Not for AI. The algorithm found that diffuse, rather than dense, plaques correlated strongly with cognitive impairment among 51 people in the cohort. ApoE4 carriers had a higher relative burden of diffuse plaques, which Bienkowski interprets as a sign of microglial failure to compact and contain Aβ aggregates. The scientists plan to develop AI-driven approaches to map other neuropathological features in more regions of the brain.
Probing Problem Proteins
AI turns out to be good at discerning how proteins fold, and which partners or substrates they bind.
While the structure of most globular proteins can be predicted from their amino acid sequence, this is not true for amyloidogenic proteins such as tau, which twist into different contortions (Oct 2021 news). To aid in atomic modeling of protein structures from cryo-EM density data, Scheres and Kiarash Jamali developed the ModelAngelo AI, a so-called graph neural network that joins cryo-EM data, amino acid sequence data, with prior knowledge about protein geometries to predict structure (Jamali et al., 2023). In structural biology, the constant development of larger graphics processing units (GPUs), which rapidly process myriad calculations in parallel, is necessary to keep up with data demands. “We developed ModelAngelo on a set of Nvidia A100 GPUs. Without cards like these, which only recently became available, our developments would not have been feasible,” Scheres said.
Regarding protein binding, Harald Steiner of Ludwig-Maximilians University Munich recently debuted a new algorithm—called comparative physicochemical profiling (CPP)— to tackle how γ-secretase recognizes its substrates. Developed by Stephan Breimann in Steiner’s lab, the algorithm went beyond sequence. It picks out physical and chemical features that distinguish substrates from non-substrates, Steiner told Alzforum. What’s more, his team validated the AI-derived γ-secretase substrates—some known, many new—at the bench with 90 percent accuracy. This research will come in handy if γ-secretase gets a fresh look as an Alzheimer's drug target.
In addition to figuring out how existing proteins fold and function, scientists are also using AI to create proteins. Case in point, today, structural and computational scientists report in Nature a generative model to design functional proteins, both monomers and oligomers (Watson et al., 2023).
Putting it All Together: Modeling and Treating Alzheimer's
AI not only is helping scientists analyze mounting data in their respective specialties, it also is helping them merge disparate types of data together to synthesize overarching models of disease. The latter is the motivation behind the dynamical systems approach proposed by Jennifer Rollo and John Hardy of University College London in a recent perspective in Neuron (Rollo et al., 2023). “There has never been greater urgency to recognize the wider complexity of dynamical systems and processes at play within a healthy brain to understand what sets it along the path of neurodegeneration,” the authors wrote. “Only with this understanding can therapeutics be found that will prevent, or at the very least, arrest, or slow the disease’s progression.”
The scientists are developing an open innovation platform, where AD researchers can combine their data and use it to test out hypotheses at the molecular, pathway, organelle, and organ levels. The platform will be devilishly complex, and AI will be critical for making it work. “In harnessing the capabilities of AI and machine learning, we can unlock new insights into the complexities of AD, minimize polypharmaceutical interventions, and pave the way for more precise and personalized approaches to diagnosis and treatment,” Rollo wrote to Alzforum (comment below). The platform will be open to universities, research institutes, industry partners, and multidisciplinary researchers.
Oskar Hanson and Jacob Vogel of Lund University in Sweden agree that multidimensional research problems such as Alzheimer's require AI. “It is in our collective interest to understand the pathway that leads from genetic polymorphism to genetic expression to protein production to biological phenotype to behavioral phenotype,” they wrote (comment below). “This is a very complex and multifaceted pathway, but the ability of many AI models to integrate complex data into low dimensional embeddings, and the ability to selectively share information across many ‘layers,’ lends itself very well to this question.”
AI and Speech: Listen and Chat
Subtle changes in speech and language are among the first symptoms in people with AD, frontotemporal dementia, and related diseases. These symptoms often go unnoticed by loved ones, even trained clinicians. AI has proven adept at picking them up (Dec 2021 conference news). For example, Winterlight Labs in Toronto uses natural language processing algorithms to detect acoustic and linguistic features and correlate them with clinical measures of disease progression (Jun 2022 conference news). Its speech-monitoring tools are exploratory outcomes in some clinical trials for AD as well as psychiatric conditions (clinicaltrials.gov). “AI can help us break down the rich information from someone’s speech into metrics that we can use to advance dementia research and develop new clinical tools,” wrote Winterlight’s Jessica Robin.
While some algorithms detect acoustic and linguistic aspects of speech, language algorithms like Generative Pre-trained Transformer 3—the behemoth behind ChatGPT—work only with text. Trained on huge amounts of it, GPT3 encodes semantic knowledge about the world through a process called embedding, which stores billions of learned patterns gleaned from the training set. Researchers at Drexel University in Philadelphia additionally trained GPT3 on snippets of text transcribed from conversations of people with or without AD. Afterward, the algorithm picked out people with AD with an accuracy of 80 percent, and predicted their MMSE scores (Agbavor and Liang, 2022). Despite being limited to text, GPT3 outperformed other algorithms that decipher acoustic features of speech.
While Winterlight and GPT3 listen to speech, other AI tools join the conversation. For example, one AI-enabled “dialogue agent” was trained on recorded video chats between interviewers and people with MCI (Tang et al., 2020). The algorithm learned how to ask questions and adapt its follow-up questions to the user’s response. In so doing, it was able to distinguish a person with normal cognition from one with early MCI, said co-author Hiroko Dodge of Massachusetts General Hospital in Boston.
Dodge heads I-CONECT, which reported a benefit of semi-structured video chats with trained conversationalists on cognitive decline in MCI (Aug 2022 conference news). The study's speech-monitoring tools indicated that these regular conversations made participants' speech more complex.
After a presentation at the 2022 AAIC, the inevitable question arose: Could robots one day replace humans in providing this social stimulus? Dodge was open to the idea at the time and, with the advent of ever more sophisticated chatbots such as ChatGPT, remains so. Still, she said that while chatbots are becoming adept at answering questions, they do not mimic the give and take of natural human conversation. “They are good at answering questions, but companionship requires more work,” she said.—Reported and written by Jessica Shugart, Homo sapiens
References
News Citations
- Massive GWAS Meta-Analysis Digs Up Trove of Alzheimer’s Genes
- From a Million Samples, GWAS Squeezes Out Seven New Alzheimer's Spots
- Are Early Harbingers of Alzheimer’s Scattered Across the Genome?
- Genetic Risk Score Combines AD GWAS Hits, Predicts Onset
- Blood Metabolome Predicts 23 Diseases, Including Dementia
- Multi-Organ MRI Unveils Myriad Connections Between Heart and Brain
- Flock of New Folds Fills in Tauopathy Family Tree
- The Rain in Spain: Move Over Higgins, AI Spots Speech Patterns
- Digital Biomarkers of FTD: How to Move from Tech Tinkering to Trials?
- A Chat Every Other Day Keeps Dementia at Bay?
Paper Citations
- Gasser U. An EU landmark for AI governance. Science. 2023 Jun 23;380(6651):1203. Epub 2023 Jun 15 PubMed.
- Clark P, Kim J, Aphinyanaphongs Y. Marketing and US Food and Drug Administration Clearance of Artificial Intelligence and Machine Learning Enabled Software in and as Medical Devices: A Systematic Review. JAMA Netw Open. 2023 Jul 3;6(7):e2321792. PubMed.
- Shah NH, Mello MM. Discrepancies Between Clearance Summaries and Marketing Materials of Software-Enabled Medical Devices Cleared by the US Food and Drug Administration. JAMA Netw Open. 2023 Jul 3;6(7):e2321753. PubMed.
- Beebe-Wang N, Celik S, Weinberger E, Sturmfels P, De Jager PL, Mostafavi S, Lee SI. Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer's disease neuropathologies. Nat Commun. 2021 Sep 10;12(1):5369. PubMed.
- Long E, Wan P, Chen Q, Lu Z, Choi J. From function to translation: Decoding genetic susceptibility to human diseases via artificial intelligence. Cell Genom. 2023 Jun 14;3(6):100320. Epub 2023 May 4 PubMed.
- Chen KM, Wong AK, Troyanskaya OG, Zhou J. A sequence-based global map of regulatory activity for deciphering human genetics. Nat Genet. 2022 Jul;54(7):940-949. Epub 2022 Jul 11 PubMed.
- Escott-Price V, Sims R, Bannister C, Harold D, Vronskaya M, Majounie E, Badarinarayan N, GERAD/PERADES, IGAP consortia, Morgan K, Passmore P, Holmes C, Powell J, Brayne C, Gill M, Mead S, Goate A, Cruchaga C, Lambert JC, van Duijn C, Maier W, Ramirez A, Holmans P, Jones L, Hardy J, Seshadri S, Schellenberg GD, Amouyel P, Williams J. Common polygenic variation enhances risk prediction for Alzheimer's disease. Brain. 2015 Dec;138(Pt 12):3673-84. Epub 2015 Oct 21 PubMed.
- Zhou X, Chen Y, Ip FC, Jiang Y, Cao H, Lv G, Zhong H, Chen J, Ye T, Chen Y, Zhang Y, Ma S, Lo RM, Tong EP, Alzheimer’s Disease Neuroimaging Initiative, Mok VC, Kwok TC, Guo Q, Mok KY, Shoai M, Hardy J, Chen L, Fu AK, Ip NY. Deep learning-based polygenic risk analysis for Alzheimer's disease prediction. Commun Med (Lond). 2023 Apr 6;3(1):49. PubMed.
- Han Y, Klinger K, Rajpal DK, Zhu C, Teeple E. Empowering the discovery of novel target-disease associations via machine learning approaches in the open targets platform. BMC Bioinformatics. 2022 Jun 16;23(1):232. PubMed.
- Pun FW, Liu BH, Long X, Leung HW, Leung GH, Mewborne QT, Gao J, Shneyderman A, Ozerov IV, Wang J, Ren F, Aliper A, Bischof E, Izumchenko E, Guan X, Zhang K, Lu B, Rothstein JD, Cudkowicz ME, Zhavoronkov A. Identification of Therapeutic Targets for Amyotrophic Lateral Sclerosis Using PandaOmics - An AI-Enabled Biological Target Discovery Platform. Front Aging Neurosci. 2022;14:914017. Epub 2022 Jun 28 PubMed.
- Steinfeldt J, Buergel T, Loock L, Kittner P, Ruyoga G, Zu Belzen JU, Sasse S, Strangalies H, Christmann L, Hollmann N, Wolf B, Ference B, Deanfield J, Landmesser U, Eils R. Neural network-based integration of polygenic and clinical information: development and validation of a prediction model for 10-year risk of major adverse cardiac events in the UK Biobank cohort. Lancet Digit Health. 2022 Feb;4(2):e84-e94. PubMed.
- Schalkamp AK, Peall KJ, Harrison NA, Sandor C. Wearable movement-tracking data identify Parkinson's disease years before clinical diagnosis. Nat Med. 2023 Aug;29(8):2048-2056. Epub 2023 Jul 3 PubMed.
- Tijms BM, Vromen EM, Mjaavatten O, Holstege H, Reus LM, vanderLee SJ, Wesenhagen K, Lorenzini L, Vermunt L, Venkatraghavan V, Tesi N, Tomassen J, denBraber A, Goossens J, Vanmechelen E, Barkhof F, Pijnenburg YA, vanderFlier WM, Teunissen CE, Berven F, Visser PJ. Large-scale cerebrospinal fluid proteomic analysis in Alzheimer's disease patients reveals five molecular subtypes with distinct genetic risk profiles. 2023 May 11 10.1101/2023.05.10.23289793 (version 1) medRxiv.
- Gogishvili D, Vromen EM, Koppes-den Hertog S, Lemstra AW, Pijnenburg YA, Visser PJ, Tijms BM, Del Campo M, Abeln S, Teunissen CE, Vermunt L. Discovery of novel CSF biomarkers to predict progression in dementia using machine learning. Sci Rep. 2023 Apr 21;13(1):6531. PubMed.
- Stephen TL, Korobkova L, Breningstall B, Nguyen K, Mehta S, Pachicano M, Jones KT, Hawes D, Cabeen RP, Bienkowski MS. Machine Learning Classification of Alzheimer's Disease Pathology Reveals Diffuse Amyloid as a Major Predictor of Cognitive Impairment in Human Hippocampal Subregions. bioRxiv. 2023 Jun 5; PubMed.
- Jamali K, Käll L, Zhang R, Brown A, Kimanius D, Scheres SH. Automated model building and protein identification in cryo-EM maps. bioRxiv. 2023 May 16; PubMed.
- Watson JL, Juergens D, Bennett NR, Trippe BL, Yim J, Eisenach HE, Ahern W, Borst AJ, Ragotte RJ, Milles LF, Wicky BI, Hanikel N, Pellock SJ, Courbet A, Sheffler W, Wang J, Venkatesh P, Sappington I, Torres SV, Lauko A, De Bortoli V, Mathieu E, Ovchinnikov S, Barzilay R, Jaakkola TS, DiMaio F, Baek M, Baker D. De novo design of protein structure and function with RFdiffusion. Nature. 2023 Jul 11; PubMed.
- Rollo J, Crawford J, Hardy J. A dynamical systems approach for multiscale synthesis of Alzheimer's pathogenesis. Neuron. 2023 Jul 19;111(14):2126-2139. Epub 2023 May 11 PubMed.
- Agbavor F, Liang H. Predicting dementia from spontaneous speech using large language models. PLOS Digit Health. 2022 Dec;1(12):e0000168. Epub 2022 Dec 22 PubMed.
- Tang F, Uchendu I, Wang F, Dodge HH, Zhou J. Scalable diagnostic screening of mild cognitive impairment using AI dialogue agent. Sci Rep. 2020 Mar 31;10(1):5732. PubMed.
External Citations
Further Reading
No Available Further Reading
Annotate
To make an annotation you must Login or Register.

Comments
Lund University
University of Pennsylvania
There is no doubt that AI is changing how we do research. It makes our work more efficient, and raises the ceiling on what we can learn. Our views will surely evolve as this technology rapidly advances, but as we see it now, there are three main situations where AI can bolster the research we are doing in neurodegenerative diseases.
The first situation, which is probably the most commonly discussed due to some high-profile successes, is the ability of AI models to potentially outperform humans in diagnostic tasks and/or discover insights from data that humans cannot or have not discovered. To reach this kind of performance, AI models often require tens or hundreds of thousands (or more) of data observations for training. An example of this might one day include predicting who will get dementia and when, using low-cost and accessible clinical markers or genetic data.
In most cases in our field, we do not yet have this much annotated data, though we can use (and actively are using) other statistical learning approaches such as machine learning and Bayesian learning to achieve greater insight in our data and better predictions. We expect AI to play a more prominent role in this domain as the size of clinical datasets continues to grow.
A second situation involves the adaptation of AI tools developed in other fields into aspects of our methodological workflow. For example, computer vision and image segmentation tools have been recently adapted to perform certain aspects of image preprocessing—a necessary step preceding data analysis—at a fraction of the speed and computational cost. While less sensational, such tools make possible the processing and quality assessment of massive datasets that otherwise would take a very long time, or even might be infeasible to process.
Another example of this situation comes from the generative tools recently making headlines, such as Dall-E 2 and Stable Diffusion. Adaptations of these can be used to generate simulated clinical images and other clinical data, which can help to facilitate data sharing, model training, and even better understanding of relationships across different types of data.
A third situation where AI tools have aided, and will continue to aid, our research agenda, is in scenarios where the architecture of AI lends itself to specific research questions. For example, it is in our collective interest to understand the pathway that leads from genetic polymorphism to genetic expression to protein production to biological phenotype to behavioral phenotype.
This is a very complex and multi-faceted pathway. The ability of many AI models to integrate complex data into low-dimensional embeddings, and the ability to selectively share information across many “layers,” lends itself very well to this question.
Finally, and maybe more controversially, would be that AI could certainty streamline the process of writing up new results into well-written scientific papers by, e.g., improving the written language. This could democratize research because non-native English-speaking persons would have a more equal chance of getting good research published in good journals. Further, AI will likely also be helpful in creating artwork and figures.
The above situations barely scratch the surface of the many ways that AI can help us perform better and more efficient research. While the simple approach of using a black box to get from existing input to desired output has potential, we think AI is more likely to be useful when implemented as a multitude of tools enhancing different nodes of an analytic processing stream. However, this field is advancing so quickly that we all must continue to be flexible, thoughtful, and creative about how to best integrate this technology as it emerges.
Columbia University Irving Medical Scool
Deploying AI methods is just beginning in the field of dementia research, in which we have a rapidly growing ecosystem of multidimensional data that is becoming increasingly complex.
Today, we are primarily repurposing datasets created for other reasons in analyses involving AI. In this context of disparate datasets that are mostly moderate in size and often have limited overlap in terms of the phenotypic features of the participants involved, AI can have a very useful role in stitching together such datasets to create a larger, shared framework that can then be explored. This can also lead to uncovering latent variables that may be important in uncovering disease-related features. However, interpreting results from AI is often difficult given that many elements of the structure that they uncover emerge from a black box.
Thus, while there is more to do with current methods and datasets, the next phase of the deployment of AI in dementia research will require (1) the construction of datasets that are designed to be analyzed by AI methods and (2) the use of “explainable AI” methods that are designed to produce more interpretable results.
This will clearly be an iterative process, but explainable AI methods are present now, and we understand that proper datasets will need to be at least an order of magnitude larger than current ones, and will need to be much more complex in terms of the dimensions of data that are available. Therefore, it is time to begin to construct these datasets.
In addition, there are more mundane tasks for which AI may be well suited and that should be strongly considered, for example, in preparing data for distribution. That is, given a curated dataset, we could use AI to (1) help write a “data descriptor text” that clearly describes the history and properties of a given dataset to facilitate its repurposing by other investigators and (2) ensure that such documents have indeed included all necessary details and that the data being distributed is in such a form that it can be readily used as directed and has the properties that are described In the data descriptor. Data distribution tasks are onerous and rarely supported by grants, so this is one area where a dedicated effort could be very fruitful in the short term.
References:
Beebe-Wang N, Celik S, Weinberger E, Sturmfels P, De Jager PL, Mostafavi S, Lee SI. Unified AI framework to uncover deep interrelationships between gene expression and Alzheimer's disease neuropathologies. Nat Commun. 2021 Sep 10;12(1):5369. PubMed.
Sasse A, Ng B, Spiro A, Tasaki S, Bennett D, Gaiteri C, De Jager PL, Chikina M, Mostafavi S. How far are we from personalized gene expression prediction using sequence-to-expression deep neural networks?. bioRxiv. 2023 Apr 26; PubMed.
Hong Kong University of Science & Technology
In our recent study (Zhou et al., 2023), we used AI for predicting the risk of developing AD based on genetic information. By employing deep learning models, specifically deep neural network (DNN), we achieved accurate classification of AD patients and have stratified individuals into distinct risk groups. This research holds immense potential for revolutionizing AD prognosis, diagnosis, and clinical research by enabling the early forecasting of AD risk with over 70 percent accuracy.
Our study highlights the broader potential of deep-learning approaches in understanding genetic contributions to diseases and the role of genetic testing in disease diagnosis. We are actively exploring the use of other such tools, including graph neural network (GNN), to incorporate additional functional genomic information such as quantitative trait locus (QTL) effects, coding/noncoding elements, and regulatory elements related to genetic variants into our study. These efforts, which we briefly discuss in the published paper, hold promise for improving the performance of risk forecasting models.
Looking ahead, we envision that with further validation and optimization in larger cohorts, our model could serve as a screening tool in routine clinical prognosis and diagnosis, improving the accuracy and confidence in AD diagnosis.
For individuals in early middle age, a genetic screen could help identify those at potential risk of developing AD, prompting them to seek genetic counsellors for appropriate follow-up.
In addition, these tools may play a crucial role in aiding drug development, enabling evaluation of drug response in people with different genetic risk levels. In the meantime, we aim to improve the disease prediction model by integrating genetic information of other AD biomarkers, such as biomedical images, and transcriptomic and proteomic profiles of CSF or blood. This would allow more accurate risk prediction and stratification of individuals according to disease progression or the involved pathogenic pathways, providing personalized information for disease intervention.
References:
Zhou X, Chen Y, Ip FC, Jiang Y, Cao H, Lv G, Zhong H, Chen J, Ye T, Chen Y, Zhang Y, Ma S, Lo RM, Tong EP, Alzheimer’s Disease Neuroimaging Initiative, Mok VC, Kwok TC, Guo Q, Mok KY, Shoai M, Hardy J, Chen L, Fu AK, Ip NY. Deep learning-based polygenic risk analysis for Alzheimer's disease prediction. Commun Med (Lond). 2023 Apr 6;3(1):49. PubMed.
Amsterdam UMC, loc. VUmc
When the ground truth is clearly known, AI will definitely push research forward in terms of efficiency, time and noise reduction. You can think of applications for cleaning and harmonizing MRI data, as well as much faster image processing. Similarly for proteomics analyses. It is hard to imagine doing my type of research without AI techniques.
That said, AI is not a magical solution that uncovers everything unknown. It remains human-made software, and so results will always be influenced by choices that a person makes, overlooked biases, and reliance on already collected data sets and interpretation, etc. So when the aim is to discover new mechanisms, I think biological intelligence comes first.
As to the specific paper mentioned in this news story (Zhou et al., 2023), it is not the AI that is "improving" the PRS per se, it is a change of outcome measure on which the PRS is based (i.e., including biological information in addition to clinical information, which may not accurately reflect underlying mechanisms). The AI just makes it easier to model many variables to then study your hypotheses.
References:
Zhou X, Chen Y, Ip FC, Jiang Y, Cao H, Lv G, Zhong H, Chen J, Ye T, Chen Y, Zhang Y, Ma S, Lo RM, Tong EP, Alzheimer’s Disease Neuroimaging Initiative, Mok VC, Kwok TC, Guo Q, Mok KY, Shoai M, Hardy J, Chen L, Fu AK, Ip NY. Deep learning-based polygenic risk analysis for Alzheimer's disease prediction. Commun Med (Lond). 2023 Apr 6;3(1):49. PubMed.
University College London
AI as a tool holds potential to advance our understanding and treatment of AD through a dynamical systems approach. Such an approach starts with a basic understanding of the system across each level—genetic, molecular, cellular, tissue, organ—and uses the minimum level of complexity necessary to capture essential details. When there is limited qualitative or quantitative data about certain interactions, different versions or parameterizations of the model are created to encompass the inherent uncertainty. These models are then employed iteratively to assess the sensitivity of the system's behaviour to uncertainties and specific network details.
We can refine models by using AI technologies that incorporate multimodal input data like genomic information, pathological data, and CT scans, along with a set of model parameters characterizing the input state of the system. These AI technologies analyse the data and parameters, map them to model output data, and compare them with corresponding experimental treatments to predict the optimal model description.
During each iteration, machine-learning algorithms modify the network description of AD. Interactions that are deemed unnecessary for modelling the dynamics or seeking interventions are excluded, while new interactions (previously unknown unknowns) that the model suggests are required but lack supporting data are introduced. This iterative process continues until the modeled behavior aligns with observed behaviour, effectively converging the model and reality.
Overall, AI facilitates the dynamical-systems approach to AD by automating the iterative refinement of our models, incorporating diverse data types, optimizing model descriptions, and identifying new interactions that require further experimental investigation.
How can the field apply this? For example, using advanced modeling techniques, our approach aims to identify the minimum set of interventions and their coordinated timing needed to restore the diseased state of the brain back to a healthy state.
In general, ML algorithms work well to reveal correlations within complex datasets, but determining causation has remained a challenge. We are therefore planning to use ML and AI approaches to efficiently search through vast parameter spaces of complex models of AD to identify configurations that accurately replicate the observed healthy state of brain tissue. By comparing the predicted behavior of the model with the pathological state, we can confirm its accuracy.
ML algorithms are also used in conjunction with the models to explore the parameter space and identify regions corresponding to both healthy and diseased states. By mapping these states to clinical manifestations of AD through imaging data, for example, we can determine the system components and interactions implicated in the pathological state.
Science, data, tools, and people are the enablers to building our proposed integrative model system and platform, and the tools include AI. In establishing an open innovation ecosystem, a global community of platform users who will utilize the AI technologies embedded into the platform will include universities, research institutes, industry partners, and multidisciplinary researchers.
Make a Comment
To make a comment you must login or register.