Induced Microglia Make Debut at Keystone Symposium
Quick Links
Ironically, microglia in a dish stole some of the limelight at “Microglia in the Brain,” a symposium held jointly with another called “Common Mechanisms of Neurodegeneration,” June 12-16 in Keystone, Colorado. No sooner had Christopher Glass cautioned in his keynote address that isolated microglia bear little resemblance to their forbears in the CNS than posters and talks about induced microglial went up—and captured the imagination of meeting-goers. While time will tell how well this new type of cultured cell mimics microglia in their natural environment, initial signs are they that have a more authentic look and feel than microglial cells currently being used in vitro. Scientists seemed buoyed by the prospect of using the new cells for questions that cannot be addressed in studies of living animals or people. “These developments are very exciting,” said Irene Knuesel, Roche Innovation Center, Basel, Switzerland. “These may not be in vivo microglia, but they provide us with a tool and an opportunity to start studying these cells in much more detail.”
Three research groups independently generated induced microglia. One group cultured progenitors together with neurons and/or astrocytes; another used a “cocktail” approach, adding just the right combination of trophic factors to cause progenitors to morph into microglia; and the third combined both approaches, which they said provides more flexible experimental platforms.
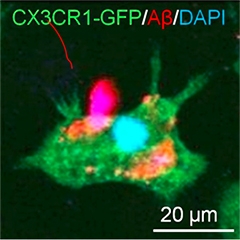
Hello microglia?
An iPSC-derived microglia-like macrophage (green) captures Aβ. [Courtesy of Florent Ginhoux and Kazuyuki Takata.]
Florent Ginhoux—known for his pioneering studies of the developmental origin of microglia—tried to recapitulate the in vivo development of microglia in an in vitro setting. In 2010, Ginhoux, who works at the Agency for Science, Technology and Research in Singapore, reported that brain-resident microglia derive from progenitors in the embryonic yolk sac (Ginhoux et al., 2010). “We basically wanted to recapitulate this primitive hematopoiesis in a dish,” Ginhoux told Alzforum. He developed a unique protocol that turns induced pluripotent stem cells (iPSCs) from healthy donors into yolk sac macrophages within a few weeks. Just like real macrophages, the cells are big and round; however, when Ginhoux cultures them together with neurons they develop microglial-like processes that reach out and touch the neurons. They also ingest synthetic Aβ fibrils (see image at left).
“It is fascinating that we can see very clear interactions between the two cell types,” said Ginhoux. While he agreed that it will be impossible to recapitulate the complexity of the brain, he plans to use iPSCs from healthy donors and people with neurodegenerative disorders, such as Parkinson’s and Alzheimer’s, to derive and co-culture neurons and glia and study interactions that may be important in disease.
Researchers working in Rudolph Jaenisch’s lab at the Whitehead Institute for Biomedical Research, Cambridge, Massachusetts, used a monoculture approach. Julien Muffat first derived primitive macrophages from human iPSCs, then used a fully defined culture medium that he developed to turn them into microglia. Muffat showed that these microglia become ramified, sporting processes with first-order branching, and that they phagocytose Aβ fibrils. Expression analysis suggests the cells’ transcriptome closely mimics that of fetal and adult microglia, rather than blood macrophages. He also said that his protocol was very robust and he has derived microglia from more than 20 different iPScell lines so far.

Cultured microglia.
Microglia derived from induced pluripotent stem cells extend cellular processes, just like microglia in the brain. [Courtesy of Julien Muffat, MIT.]
Muffat was able to shift that transcriptional signature closer to that seen in microglia in the brain by culturing the induced microglia in three dimensions with neurons and astrocytes. In the three-dimensional cultures, the microglia develop long processes that extend and retract (see image at left). In a laser-ablation assay, where researchers make a tiny wound by burning a hole through tissue, the induced microglia extended toward, and clustered around, sites of injury, much like microglia do around amyloid plaques in the brain. In cerebral organoids the induced microglia cells appear to “tile” the way they do in vivo in the brain.
Muffat is now collaborating with researchers in Rudy Tanzi’s lab at Massachusetts General Hospital, Charlestown, to embed these cells in three-dimensional neuronal cultures that generate amyloid plaques and neurofibrillary tangles, the hallmarks of AD (see Oct 2014 news).
Also taking a more direct stab at developing induced microglia, Edsel Abud in Mathew Blurton-Jones’ lab at the University of California, Irvine, shied away from using neurons or other cells. “The main difference with our approach is that it allows us to study microglia without the need for complex co-culture systems,” said Blurton-Jones. “One potential advantage is that it will be easier to conduct high-throughput quantitative screening.” Some pharma researchers at the meeting lamented that they cannot carry out high-throughput screens on microglia using tissue slice preparations, but thought this approach might prove valuable for translational research.
Abud built on years of studies on mouse microglial ontology that highlight not only where microglia originate, but also what they need to thrive. He told Alzforum that he tried to identify from the literature the components that seemed necessary for microglia to survive, mature, and stay healthy, and then tested those components empirically. “We thought we’d try a reductionist approach and find the soluble factors that would be needed to derive and maintain microglia,” he said. Abud modified a protocol used to derive primitive hematopoietic progenitors from iPS lines, and then guided those cells toward a microglia-like phenotype with a mixture of different factors.
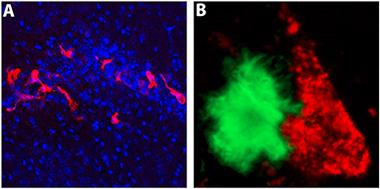
iPS-microglia in the brain.
When transplanted into the hippocampi of immune-deficient 5xfAD mice, microglia derived from induced pluripotent stem cells (red) migrate and interact with Aβ plaques (green). [Courtesy of Matt Blurton-Jones, UC Irvine.]
These iPS microglia (iPS-MG) express typical microglial markers, including P2Y12R, TREM2, DAP12, and CX3CR1. Based on co-expression of P2Y12R and TREM2, these cells are more than 96 percent pure, said Blurton-Jones. Abud and colleagues used three-dimensional principle component analysis, a method to pull out the most significant trends in a data set, to compare whole-genome transcriptional profiles of the iPS-MG against those of other cells. The iPS-MG profiles matched closely with those of adult and fetal microglia but diverged from those of blood monocytes.
Blurton-Jones and colleagues are studying how the iPS-MG respond to Aβ plaques and neurofibrillary tangles, the hallmarks of AD pathology. On his Keystone poster, Abud reported that the cells phagocytose fibrillar Aβ, and upregulate several AD-associated genes in the process, including TREM2, ApoE, ABCA7, and PICALM. Clusterin stood out for its 35-fold elevation in expression. In collaboration with Rakez Kayed at University of Texas Medical Branch, Galveston, Abud found that the cells also take up tau oligomers isolated from human brain. Working with Karen Gylys from the University of California, Los Angeles, the group also found that iPS-MG could phagocytose human brain-derived synaptosomes and that blocking components of complement receptor 3 (CR3) could impair this process. Researchers led by Beth Stevens at Children’s Hospital, Boston, have shown the microglia prune synapses in the adult brain in a CR3-dependent manner, and that this can be exacerbated by Aβ (see Apr 2016 news).
Ginhoux noted that having these different ways to make induced microglia is good for the field. “This is how we progress in science. It should give others the opportunity to build and improve on these approaches,” he said.
Keynote speaker Glass, of the University of California, San Diego, said that going from iPS or embryonic stem cell to in vitro microglia represents a huge advance for the field. “It seems that these researchers have made substantial progress,” he told Alzforum. But he cautioned that in vitro cells may not always tell us what happens in vivo. On that score, Blurton-Jones and colleagues have transplanted induced microglia into the hippocampus of immune-deficient 5xFAD mice (Marsh et al., 2016), where they began to migrate toward and interact with Aβ plaques (see image above). Glass also emphasized the need to study human microglia in a human brain context, and suggested organoids might be the way to go. Muffat and colleagues are partway there with their three-dimensional culture systems.
All three groups plan to study how genetic variation affects their induced cells. “We can now try to figure out how variants in genes such as TREM2 affect the different cell types in the brain,” said Ginhoux. Blurton-Jones and colleagues have already begun to culture induced microglia from people carrying the R47H and T66M TREM2 mutations, which are risk alleles for Alzheimer’s and frontotemporal dementia, respectively.
The researchers agreed that these cell lines can help the field get past a niggling problem in studying risk alleles identified by genome-wide association studies. As emphasized by Glass and others at the meeting, genetic heterogeneity in the population presents a major hurdle in interpreting the effect of GWAS hits on disease pathology. To model this, Ginhoux, Muffat, and Blurton-Jones plan to use CRISPR technology to introduce genetic variants into stable microglial lines. In Keystone, Muffat reported using CRISPR to generate induced microglial with the presenilin 1 A246E mutation and with TREM2 knocked out. “Using isogenic lines will help us account for background genetic variation that might confound analysis,” noted Blurton-Jones.—Tom Fagan
References
News Citations
- Alzheimer’s in a Dish? Aβ Stokes Tau Pathology in Third Dimension
- Paper Alert: Microglia Mediate Synaptic Loss in Early Alzheimer’s Disease
Alzpedia Citations
Research Models Citations
Paper Citations
- Ginhoux F, Greter M, Leboeuf M, Nandi S, See P, Gokhan S, Mehler MF, Conway SJ, Ng LG, Stanley ER, Samokhvalov IM, Merad M. Fate mapping analysis reveals that adult microglia derive from primitive macrophages. Science. 2010 Nov 5;330(6005):841-5. PubMed.
- Marsh SE, Abud EM, Lakatos A, Karimzadeh A, Yeung ST, Davtyan H, Fote GM, Lau L, Weinger JG, Lane TE, Inlay MA, Poon WW, Blurton-Jones M. The adaptive immune system restrains Alzheimer's disease pathogenesis by modulating microglial function. Proc Natl Acad Sci U S A. 2016 Mar 1;113(9):E1316-25. Epub 2016 Feb 16 PubMed.
Other Citations
Further Reading
No Available Further Reading
Annotate
To make an annotation you must Login or Register.

Comments
No Available Comments
Make a Comment
To make a comment you must login or register.